|
CooA
CooA is a heme-containing transcription factor that responds to the presence of carbon monoxide. This protein forms homodimers and is a homolog of cAMP receptor protein. CooA regulates the expression of carbon monoxide dehydrogenase, an enzyme that catalyzes the oxidation of CO to CO2. The most well-studied CooA homolog comes from ''Rhodospirillum rubrum ''Rhodospirillum rubrum'' (''R. rubrum'') is a Gram-negative, pink-coloured bacterium, with a size of 800 to 1000 nanometers. It is a facultative anaerobe, thus capable of using oxygen for aerobic respiration under aerobic conditions, or an alte ...'' (''RrCooA),'' but the CooA homolog from '' Carboxydothermus hydrogenoformans'' (''Ch''CooA) has been studied as well. The main distinction between these two CooA homologs is the ferric heme coordination. For ''Rr''CooA, the ferric heme iron is bound to a cysteine and the amine of the N-terminal proline, while, in the ferrous state, a ligand switch occurs where a nearby histidine d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
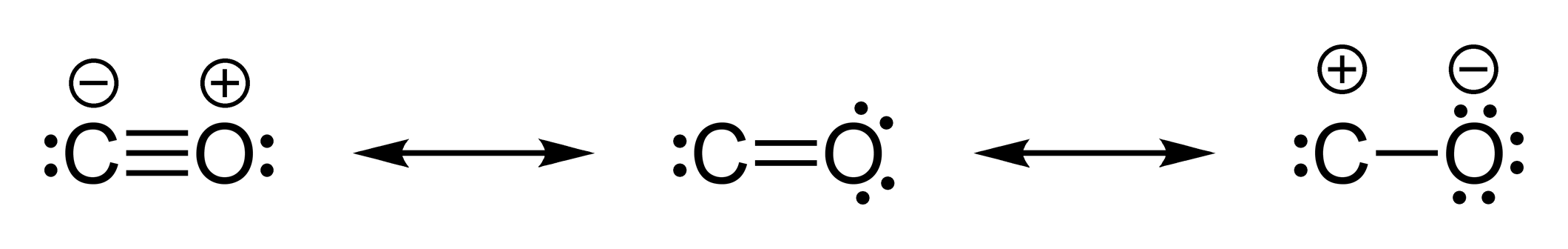
Carbon Monoxide
Carbon monoxide (chemical formula CO) is a colorless, poisonous, odorless, tasteless, flammable gas that is slightly less dense than air. Carbon monoxide consists of one carbon atom and one oxygen atom connected by a triple bond. It is the simplest molecule of the oxocarbon family. In coordination complexes the carbon monoxide ligand is called carbonyl. It is a key ingredient in many processes in industrial chemistry. The most common source of carbon monoxide is the partial combustion of carbon-containing compounds, when insufficient oxygen or heat is present to produce carbon dioxide. There are also numerous environmental and biological sources that generate and emit a significant amount of carbon monoxide. It is important in the production of many compounds, including drugs, fragrances, and fuels. Upon emission into the atmosphere, carbon monoxide affects several processes that contribute to climate change. Carbon monoxide has important biological roles across phylogenetic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heme
Heme, or haem (pronounced / hi:m/ ), is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver. In biochemical terms, heme is a coordination complex "consisting of an iron ion coordinated to a porphyrin acting as a tetradentate ligand, and to one or two axial ligands." The definition is loose, and many depictions omit the axial ligands. Among the metalloporphyrins deployed by metalloproteins as prosthetic groups, heme is one of the most widely used and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigment in blood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and endothelial nitric oxide synthase. The word ''haem'' is derived from Greek ''haima'' meaning "blood". Function Hemoproteins have diverse biological functions incl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CAMP Receptor Protein
cAMP receptor protein (CRP; also known as catabolite activator protein, CAP) is a regulatory protein in bacteria. CRP protein binds cAMP, which causes a conformational change that allows CRP to bind tightly to a specific DNA site in the promoters of the genes it controls. CRP then activates transcription through direct protein–protein interactions with RNA polymerase. The genes regulated by CRP are mostly involved in energy metabolism, such as galactose, citrate, or the PEP group translocation system. In ''Escherichia coli'', cyclic AMP receptor protein (CRP) can regulate the transcription of more than 100 genes. The signal to activate CRP is the binding of cyclic AMP. Binding of cAMP to CRP leads to a long-distance signal transduction from the N-terminal cAMP-binding domain to the C-terminal domain of the protein, which is responsible for interaction with specific sequences of DNA. At "Class I" CRP-dependent promoters, CRP binds to a DNA site located upstream of core promo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbon Monoxide Dehydrogenase
In enzymology, carbon monoxide dehydrogenase (CODH) () is an enzyme that catalyzes the chemical reaction :CO + H2O + A \rightleftharpoons CO2 + AH2 The chemical process catalyzed by carbon monoxide dehydrogenase is similar to the water-gas shift reaction. The 3 substrates of this enzyme are CO, H2O, and A, whereas its two products are CO2 and AH2. A variety of electron donors/receivers (Shown as "A" and "AH2" in the reaction equation above) are observed in micro-organisms which utilize CODH. Several examples of electron transfer cofactors has been proposed, including Ferredoxin, NADP+/NADPH and flavoprotein complexes like flavin adenine dinucleotide (FAD) as well as hydrogenases. CODHs support the metabolisms of diverse prokaryotes, including methanogens, aerobic carboxidotrophs, acetogens, sulfate-reducers, and hydrogenogenic bacteria. The bidirectional reaction catalyzed by CODH plays a role in the carbon cycle allowing organisms to both make use of CO as a source of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhodospirillum Rubrum
''Rhodospirillum rubrum'' (''R. rubrum'') is a Gram-negative, pink-coloured bacterium, with a size of 800 to 1000 nanometers. It is a facultative anaerobe, thus capable of using oxygen for aerobic respiration under aerobic conditions, or an alternative terminal electron acceptor for anaerobic respiration under anaerobic conditions. Alternative terminal electron acceptors for ''R. rubrum'' include dimethyl sulfoxide or trimethylamine oxide. Under aerobic growth photosynthesis is genetically suppressed and ''R. rubrum'' is then colorless. After the exhaustion of oxygen, ''R. rubrum'' immediately starts the production of photosynthesis apparatus including membrane proteins, bacteriochlorophylls and carotenoids, i.e. the bacterium becomes photosynthesis active. The repression mechanism for the photosynthesis is poorly understood. The photosynthesis of ''R. rubrum'' differs from that of plants as it possesses not chlorophyll a, but bacteriochlorophylls. While bacteriochlorophyll can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carboxydothermus Hydrogenoformans
''Carboxydothermus hydrogenoformans'' is an extremely thermophilic anaerobic Gram-positive bacterium that has the interesting property of producing hydrogen as a waste product while feeding on carbon monoxide and water. It also forms endospores. It was isolated from a hot spring on the Russian volcanic island of Kunashir by Svetlichny et al. in 1991. Its complete genome was sequenced in 2005 by a team of scientists of the Institute for Genomic Research (TIGR) According to TIGR evolutionary biologist Jonathan Eisen, ''"C. hydrogenoformans is one of the fastest-growing microbes that can convert water and carbon monoxide to hydrogen."'' The microbe owes this to the fact that it has at least five different forms of carbon monoxide dehydrogenase In enzymology, carbon monoxide dehydrogenase (CODH) () is an enzyme that catalyzes the chemical reaction :CO + H2O + A \rightleftharpoons CO2 + AH2 The chemical process catalyzed by carbon monoxide dehydrogenase is similar to the water- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein familie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA po ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |