|
C Cap
The term C cap (C-cap, Ccap) describes an amino acid in a particular position within a protein or polypeptide.{{cite journal, last=Leader, first=DP, author2=Milner-White EJ , title=The structure of the ends of helices in globular proteins, journal=Proteins, year=2011, volume=79, issue=3, pages=1010–1019, doi= 10.1002/prot.22942, pmid=21287629, s2cid=22240314 The C cap residue of an alpha helix is the last amino acid residue at the C terminus of the helix. More precisely, it is defined as the last residue (i) whose NH group is hydrogen-bonded to the CO group of residue i-4 (or sometimes residue i-3). Because of this it is sometimes also described as the residue following the helix. Certain motifs occur commonly at or around the C cap, notably the Schellman loop and the niche (protein structural motif). The N cap The term N cap (N-cap, Ncap) describes an amino acid in a particular position within a protein or polypeptide.{{cite journal, last=Leader, first=DP, author2=Milner-White ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypeptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C Terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often conta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen-bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted , where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the second-row elements nitrogen (N), oxygen (O), and fluorine (F). Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). The energy of a hydrogen bond depends on the geometry, the environment, and the nature of the specific donor and acceptor atoms and can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully coval ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Schellman Loop
Schellman loops (also called Schellman motifs or paperclips) are commonly occurring structural features of proteins and polypeptides. Each has six amino acid residues (labelled residues ''i'' to ''i''+5) with two specific inter-mainchain hydrogen bonds (as in lower figure, i) and a characteristic main chain dihedral angle conformation. The CO group of residue ''i'' is hydrogen-bonded to the NH of residue ''i''+5 (colored orange in upper figure), and the CO group of residue ''i''+1 is hydrogen-bonded to the NH of residue ''i''+4 (beta turn, colored purple). Residues ''i''+1, ''i''+2, and ''i''+3 have negative φ (phi) angle values and the phi value of residue ''i''+4 is positive. Schellman loops incorporate a three amino acid residue RL nest (protein structural motif), in which three mainchain NH groups (from Schellman loop residues ''i''+3 to ''i''+5) form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
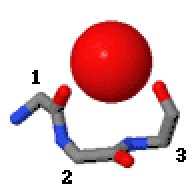
Niche (protein Structural Motif)
In the area of protein structural motifs, niches are three or four amino acid residue features in which main-chain CO groups are bridged by positively charged or δ+ groups. The δ+ groups include groups with two hydrogen bond donor atoms such as NH2 groups and water molecules. In typical proteins, 7% of amino acid residues belong to niches bound to a δ+ group, while another 7% have the conformation but no single cationic bridging group is detected. Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''. A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''. A niche occurs commonly at the C-terminus of α-helices ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N Cap
The term N cap (N-cap, Ncap) describes an amino acid in a particular position within a protein or polypeptide.{{cite journal, last=Leader, first=DP, author2=Milner-White EJ , title=The structure of the ends of helices in globular proteins, journal=Proteins, year=2011, volume=79, issue=3, pages=1010–1019, doi= 10.1002/prot.22942, pmid=21287629, s2cid=22240314 The N cap residue of an alpha helix is the first amino acid residue at the N terminus of the helix. More precisely, it is defined as the first residue (i) whose CO group is hydrogen-bonded to the NH group of residue i+4 (or sometimes residue i+3). Because of this it is sometimes also described as the residue prior to the helix. Capping motifs are those often found at the N cap. Asx turns, ST turns, and asx motifs are often found at such situations, with the asx or serine or threonine residue at the N cap. The C cap The term C cap (C-cap, Ccap) describes an amino acid in a particular position within a protein or polypeptide.{ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |