|
COX4I2
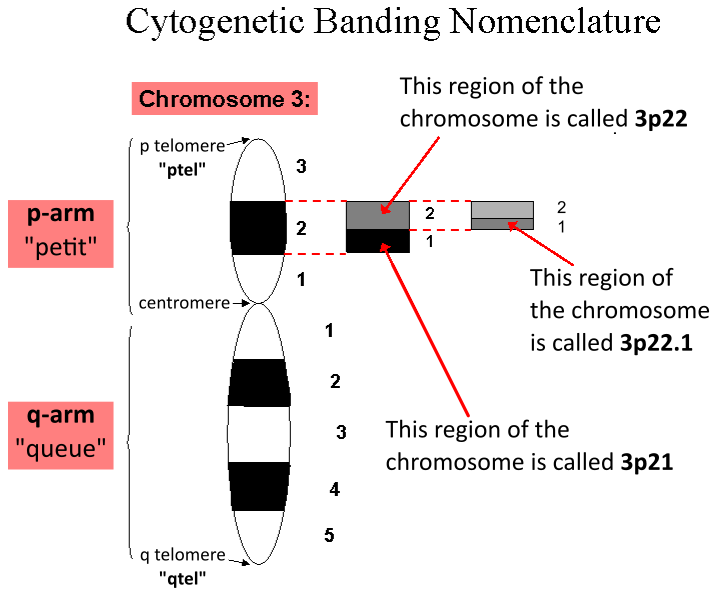
Cytochrome c oxidase subunit 4 isoform 2, mitochondrial is an enzyme that in humans is encoded by the ''COX4I2'' gene. COX4I2 is a nuclear-encoded isoform of cytochrome c oxidase (COX) subunit 4. Cytochrome c oxidase ( complex IV) is a multi-subunit enzyme complex that couples the transfer of electrons from cytochrome c to molecular oxygen and contributes to a proton electrochemical gradient across the inner mitochondrial membrane, acting as the terminal enzyme of the mitochondrial respiratory chain. Mutations in ''COX4I2'' have been associated with exocrine pancreatic insufficiency, dyserythropoietic anemia, and calvarial hyperostosis (EPIDACH). Structure ''COX4I2'' is located on the q arm of chromosome 20 in position 11.21 and has 6 exons. The ''COX4I2'' gene produces a 20 kDa protein composed of 171 amino acids. The protein encoded by ''COX4I2'' belongs to the cytochrome c oxidase IV family. ''COX4I2'' has a transit peptide domain and a disulfide bond amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
COX4I1
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1) is an enzyme that in humans is encoded by the ''COX4I1'' gene. COX4I1 is a nuclear-encoded isoform of cytochrome c oxidase (COX) subunit 4. Cytochrome c oxidase (complex IV) is a multi-subunit enzyme complex that couples the transfer of electrons from cytochrome c to molecular oxygen and contributes to a proton electrochemical gradient across the inner mitochondrial membrane, acting as the terminal enzyme of the mitochondrial respiratory chain. Antibodies against COX4 can be used to identify the inner membrane of mitochondria in immunfluorescence studies. Mutations in COX4I1 have been associated with COX deficiency and Fanconi anemia. Structure ''COX4I1'' is located on the q arm of chromosome 16 in position 24.1 and has 6 exons. The ''COX4I1'' gene produces a 9.3 kDa protein composed of 83 amino acids. ''COX4I1'' is expressed ubiquitously. The protein encoded by ''COX4I1'' belongs to the cytochrome c oxidase IV fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C Oxidase
The enzyme cytochrome c oxidase or Complex IV, (was , now reclassified as a translocasEC 7.1.1.9 is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes. It is the last enzyme in the respiratory electron transport chain of cells located in the membrane. It receives an electron from each of four cytochrome c molecules and transfers them to one oxygen molecule and four protons, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton electrochemical potential, which the ATP synthase then uses to synthesize ATP. Structure The complex The complex is a large integral membrane protein composed of several metal prosthetic sites and 14 protein subunits in mammals. In mammals, eleven subunits are nuclear in origin, and three are synthesized in the mitochondria. The complex contains two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C Oxidase
The enzyme cytochrome c oxidase or Complex IV, (was , now reclassified as a translocasEC 7.1.1.9 is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes. It is the last enzyme in the respiratory electron transport chain of cells located in the membrane. It receives an electron from each of four cytochrome c molecules and transfers them to one oxygen molecule and four protons, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton electrochemical potential, which the ATP synthase then uses to synthesize ATP. Structure The complex The complex is a large integral membrane protein composed of several metal prosthetic sites and 14 protein subunits in mammals. In mammals, eleven subunits are nuclear in origin, and three are synthesized in the mitochondria. The complex contains two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heteromer
A heteromer is something that consists of different parts; the antonym of homomeric. Examples are: Biology * Spinal neurons that pass over to the opposite side of the spinal cord. * A protein complex that contains two or more different polypeptides. Pharmacology * Ligand-gated ion channels such as the nicotinic acetylcholine receptor and GABAA receptor are composed of five subunits arranged around a central pore that opens to allow ions to pass through. There are many different subunits available that can come together in a wide variety of combinations to form different subtypes of the ion channel. Sometimes the channel can be made from only one type of subunit, such as the α7 nicotinic receptor, which is made up from five α7 subunits, and so is homomerrather than a heteromer, but more commonly several different types of subunit will come together to form a heteromeric complex (e.g., the α4β2 nicotinic receptor, which is made up from two α4 subunits and three β2 subunit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial Respiratory Chain
An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electrons that transferred from NADH and FADH2 to the ETC involves 4 multi-subunit large enzymes complexes and 2 mobile electron carriers. Many of the enzymes in the electron transport chain are membrane-bound. The flow of electrons through the electron transport chain is an exergonic process. The energy from the redox reactions creates an electrochemical proton gradient that drives the synthesis of adenosine triphosphate (ATP). In aerobic respiration, the flow of electrons terminates with molecular oxygen as the final electron acceptor. In anaerobic respiration, other electron acceptors are used, such as sulfate. In an electron transport chain, the redox r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypoxia (medical)
Hypoxia is a condition in which the body or a region of the body is deprived of adequate oxygen supply at the tissue level. Hypoxia may be classified as either '' generalized'', affecting the whole body, or ''local'', affecting a region of the body. Although hypoxia is often a pathological condition, variations in arterial oxygen concentrations can be part of the normal physiology, for example, during strenuous physical exercise.. Hypoxia differs from hypoxemia and anoxemia, in that hypoxia refers to a state in which oxygen present in a tissue or the whole body is insufficient, whereas hypoxemia and anoxemia refer specifically to states that have low or no oxygen in the blood. Hypoxia in which there is complete absence of oxygen supply is referred to as anoxia. Hypoxia can be due to external causes, when the breathing gas is hypoxic, or internal causes, such as reduced effectiveness of gas transfer in the lungs, reduced capacity of the blood to carry oxygen, compromised general ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, prompt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glutamic Acid
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synthesize enough for its use. It is also the most abundant excitatory neurotransmitter in the vertebrate nervous system. It serves as the precursor for the synthesis of the inhibitory gamma-aminobutyric acid (GABA) in GABA-ergic neurons. Its molecular formula is . Glutamic acid exists in three optically isomeric forms; the dextrorotatory -form is usually obtained by hydrolysis of gluten or from the waste waters of beet-sugar manufacture or by fermentation.Webster's Third New International Dictionary of the English Language Unabridged, Third Edition, 1971. Its molecular structure could be idealized as HOOC−CH()−()2−COOH, with two carboxyl groups −COOH and one amino group −. However, in the solid state and mildly acidic water solutio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bond
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251& ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to translo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |