|
CDNAs
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms ( SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase I
DNA polymerase I (or Pol I) is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase (and the first known of any kind of polymerase). It was initially characterized in '' E. coli'' and is ubiquitous in prokaryotes. In ''E. coli'' and many other bacteria, the gene that encodes Pol I is known as ''polA''. The ''E. coli'' Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA. Discovery In 1956, Arthur Kornberg and colleagues discovered Pol I by using ''Escherichia coli'' (''E. coli'') extracts to develop a DNA synthesis assay. The scient ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important list of RNAs, types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, Extracellular RNA, exRNAs, scaRNAs and the long noncoding RNA, long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent Transcriptomics, transcriptomic and Bioinformatics, bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create thre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Random Hexamer
A random hexamer or random hexonucleotides are for various PCR applications such as rolling circle amplification Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genom ... to prime the DNA. They are oligonucleotide sequences of 6 bases which are synthesised entirely randomly to give a numerous range of sequences that have the potential to anneal at many random points on a DNA sequence and act as a primer to commence first strand cDNA synthesis. References {{reflist Polymerase chain reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thymine
Thymine () ( symbol T or Thy) is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. In RNA, thymine is replaced by the nucleobase uracil. Thymine was first isolated in 1893 by Albrecht Kossel and Albert Neumann from calf thymus glands, hence its name. Derivation As its alternate name (5-methyluracil) suggests, thymine may be derived by methylation of uracil at the 5th carbon. In RNA, thymine is replaced with uracil in most cases. In DNA, thymine (T) binds to adenine (A) via two hydrogen bonds, thereby stabilizing the nucleic acid structures. Thymine combined with deoxyribose creates the nucleoside deoxythymidine, which is synonymous with the term thymidine. Thymidine can be phosphorylated with up to three phosphoric acid groups, producing dTMP (deoxythymidine monophosphate), dTDP, or dTTP (for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
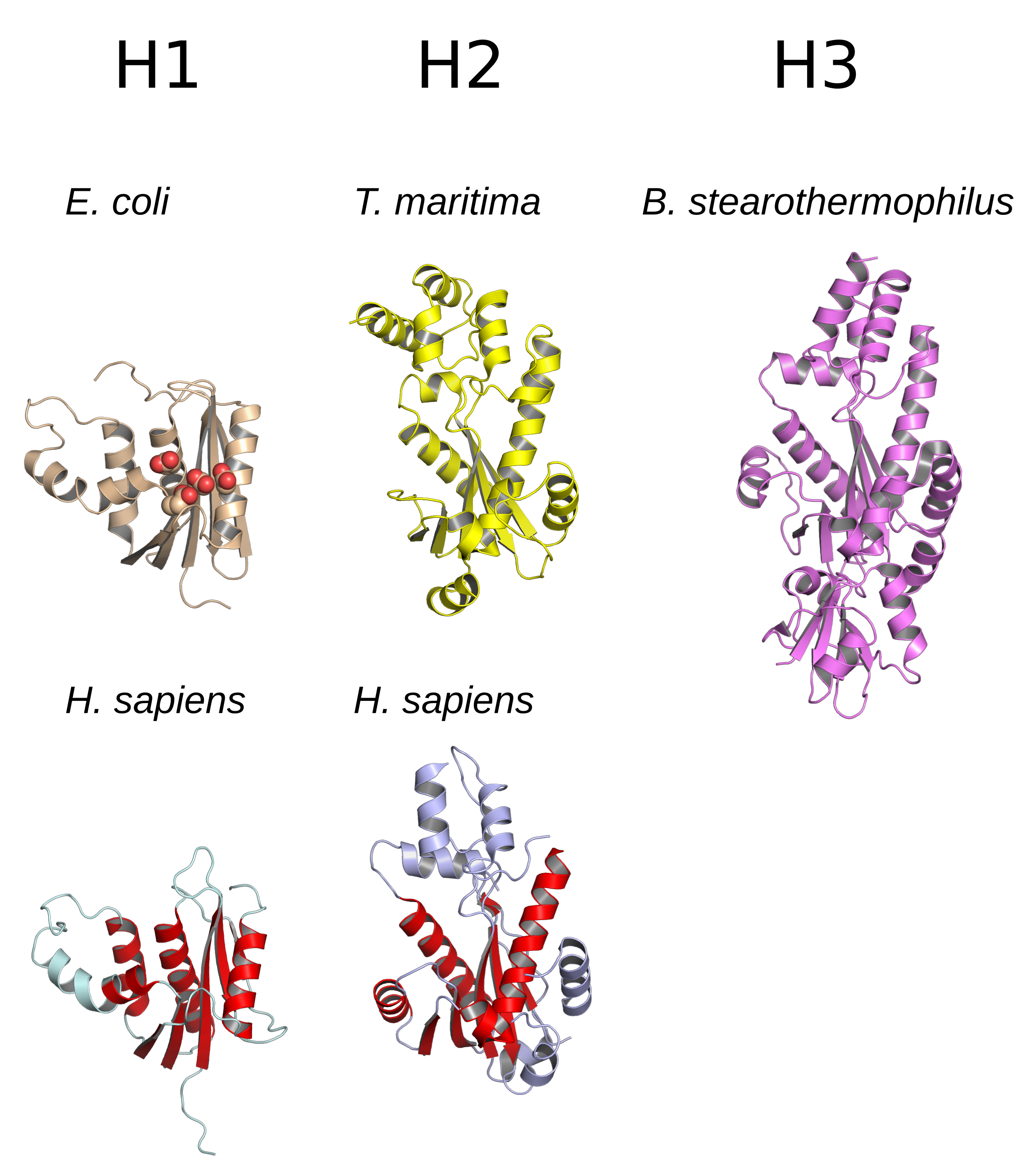
Ribonuclease H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for vira ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme ( RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysis
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular biology, biochemistry, and cell biology laboratories, cell cultures may be subjected to lysis in the process of purifying their components, as in protein purification, DNA extraction, RNA extraction, or in purifying organelles. Many species of bacteria are subject to lysis by the enzyme lysozyme, found in animal saliva, egg white, and other secretions. Phage lytic enzymes (lysins) produced during bacteriophage infection are responsible for the ability of these viruses to lyse bacterial cells. Penicillin and related β-lactam antibiotics cause the death of bacteria through enzyme-mediated lysis that occurs after the drug causes the bacterium to form a defective cell wall. If the cell wall is completely lost and the penicillin was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Extraction
RNA extraction is the purification of RNA from biological samples. This procedure is complicated by the ubiquitous presence of ribonuclease enzymes in cells and tissues, which can rapidly degrade RNA. Several methods are used in molecular biology to isolate RNA from samples, the most common of these is guanidinium thiocyanate-phenol-chloroform extraction. The filter paper based lysis and elution method features high throughput capacity. RNA extraction in liquid nitrogen, commonly using a mortar and pestle (or specialized steel devices known as tissue pulverizers) is also useful in preventing ribonuclease activity. RNase contamination The extraction of RNA in molecular biology experiments is greatly complicated by the presence of ubiquitous and hardy RNases that degrade RNA samples. Certain RNases can be extremely hardy and inactivating them is difficult compared to neutralizing DNases. In addition to the cellular RNases that are released there are several RNases that are present ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |