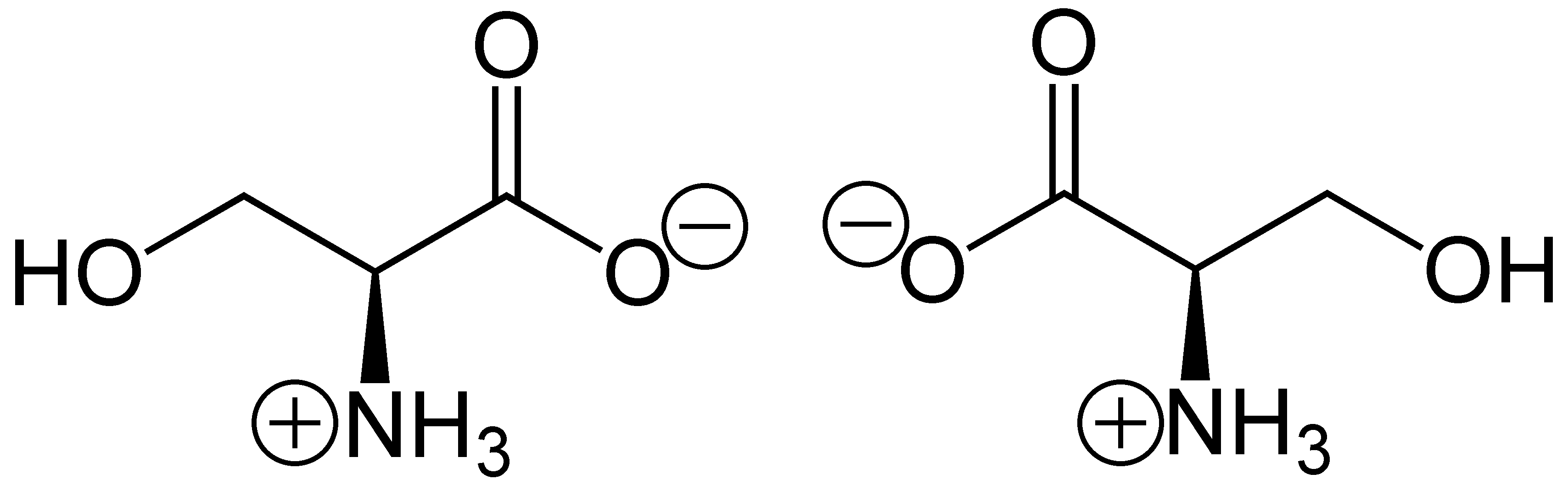|
C14orf102
Chromosome 14 open reading frame 102 (accession: AK294958; alias: C14orf102, FLJ14051, FLJ10008, FLJ52106) is a 3810bp protein-encoding gene that is highly conserved among its non-distant orthologs. It contains 20 introns and 8 different RNAs - 7 splice variants and 1 unspliced form - and is located on the reverse strand of chromosome 14 (14q32.11). The protein encoded by this gene (accession: BAG58032) belongs to the UPF0614 family of Up-frameshift proteins and has a molecular weight of 132.417 kDa and isoelectric point of 7.88. It is expected to have a protein binding function and localization in the cytoplasm. Expression According to EST profiles, C14orf102 is most highly expressed in the esophagus (98 TPM) and trachea (76), and is further expressed in multiple different tissues. However, the AceView summary suggests that this gene is expressed most highly in the brain, followed by decreased expression in 81 separate tissues. This is inferred in the reconstruction of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carrier Protein
A membrane transport protein (or simply transporter) is a membrane protein involved in the movement of ions, small molecules, and macromolecules, such as another protein, across a biological membrane. Transport proteins are integral transmembrane proteins; that is they exist permanently within and span the membrane across which they transport substances. The proteins may assist in the movement of substances by facilitated diffusion or active transport. The two main types of proteins involved in such transport are broadly categorized as either ''channels'' or ''carriers''. The solute carriers and atypical SLCs are secondary active or facilitative transporters in humans. Collectively membrane transporters and channels are known as the transportome. Transportomes govern cellular influx and efflux of not only ions and nutrients but drugs as well. Difference between channels and carriers A carrier is not open simultaneously to both the extracellular and intracellular environments. Ei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to an mR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNPS1
RNA-binding protein with serine-rich domain 1 is a protein that in humans is encoded by the ''RNPS1'' gene. Function This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junction complex, involved in both mRNA nuclear export and mRNA surveillance. mRNA surveillance detects exported mRNAs with truncated open reading frames and initiates nonsense-mediated mRNA decay (NMD). When translation ends upstream from the last exon-exon junction, this triggers NMD to degrade mRNAs containing premature stop codons. This protein binds to the mRNA and remains bound after nuclear export, acting as a nucleocytoplasmic shuttling protein. This protein contains many serine residues. Two splice variants have been found for this gene; both variants encode the same protein. Interactions RNPS1 has been shown to interact with SART3 and Pinin Pinin is a protein that in humans is encoded by the ''PNN'' gene. Interactions Pinin has been shown to interact with Ke ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Codons
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Efforts ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences. In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics. Methanogenesis Methanogenesis, the process th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Protein phosphorylation often activates (or deactivates) many enzymes. Glucose Phosphorylation of sugars is often the first stage in their catabolism. Phosphorylation allows cells to accumulate sugars because the phosphate group prevents the molecules from diffusing back across their transporter. Phosphorylation of glucose is a key reaction in sugar metabolism. The chemical equation for the conversion of D-glucose to D-glucose-6-phosphate in the first step of glycolysis is given by :D-glucose + ATP → D-glucose-6-phosphate + ADP : ΔG° = −16.7 kJ/mol (° indicates measurement at standard condition) Hepatic cells are freely permeable to glucose, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poly-lysine
Polylysine refers to several types of lysine homopolymers, which may differ from each other in terms of stereochemistry (D/L; the L form is natural and usually assumed) and link position (α/ε). Of these types, only ε-poly-L-lysine is produced naturally. Chemical structure The precursor amino acid lysine contains two amino groups, one at the α-carbon and one at the ε-carbon. Either can be the location of polymerization, resulting in α-polylysine or ε-polylysine. Polylysine is a homopolypeptide belonging to the group of cationic polymers: at pH 7, polylysine contains a positively charged hydrophilic amino group. α-Polylysine is a synthetic polymer, which can be composed of either L-lysine or D-lysine. "L" and "D" refer to the chirality at lysine's central carbon. This results in poly-L-lysine (PLL) and poly-D-lysine (PDL) respectively. ε-Polylysine (ε-poly-L-lysine, EPL) is typically produced as a homopolypeptide of approximately 25–30 L-lysine residues. According ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain lysyl ((CH2)4NH2), classifying it as a basic, charged (at physiological pH), aliphatic amino acid. It is encoded by the codons AAA and AAG. Like almost all other amino acids, the α-carbon is chiral and lysine may refer to either enantiomer or a racemic mixture of both. For the purpose of this article, lysine will refer to the biologically active enantiomer L-lysine, where the α-carbon is in the ''S'' configuration. The human body cannot synthesize lysine. It is essential in humans and must therefore be obtained from the diet. In organisms that synthesise lysine, two main biosynthetic pathways exist, the diaminopimelate and α-aminoadipate pathways, which employ distinct e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |