|
Baum–Welch Algorithm
In electrical engineering, statistical computing and bioinformatics, the Baum–Welch algorithm is a special case of the expectation–maximization algorithm used to find the unknown parameters of a hidden Markov model (HMM). It makes use of the forward-backward algorithm to compute the statistics for the expectation step. History The Baum–Welch algorithm was named after its inventors Leonard E. Baum and Lloyd R. Welch. The algorithm and the Hidden Markov models were first described in a series of articles by Baum and his peers at the IDA Center for Communications Research, Princeton in the late 1960s and early 1970s. One of the first major applications of HMMs was to the field of speech processing. In the 1980s, HMMs were emerging as a useful tool in the analysis of biological systems and information, and in particular genetic information. They have since become an important tool in the probabilistic modeling of genomic sequences. Description A hidden Markov model describes th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electrical Engineering
Electrical engineering is an engineering discipline concerned with the study, design, and application of equipment, devices, and systems which use electricity, electronics, and electromagnetism. It emerged as an identifiable occupation in the latter half of the 19th century after commercialization of the electric telegraph, the telephone, and electrical power generation, distribution, and use. Electrical engineering is now divided into a wide range of different fields, including computer engineering, systems engineering, power engineering, telecommunications, radio-frequency engineering, signal processing, instrumentation, photovoltaic cells, electronics, and optics and photonics. Many of these disciplines overlap with other engineering branches, spanning a huge number of specializations including hardware engineering, power electronics, electromagnetics and waves, microwave engineering, nanotechnology, electrochemistry, renewable energies, mechatronics/control, and electrical m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GLIMMER
In bioinformatics, GLIMMER (Gene Locator and Interpolated Markov ModelER) is used to find genes in prokaryotic DNA. "It is effective at finding genes in bacteria, archea, viruses, typically finding 98-99% of all relatively long protein coding genes". GLIMMER was the first system that used the interpolated Markov model to identify coding regions. The GLIMMER software is open source and is maintained by Steven Salzberg, Art Delcher, and their colleagues at the ''Center for Computational Biology'' at Johns Hopkins University. The original GLIMMER algorithms and software were designed by Art Delcher, Simon Kasif and Steven Salzberg and applied to bacterial genome annotation in collaboration with Owen White. Versions GLIMMER 1.0 First Version of GLIMMER "i.e., GLIMMER 1.0" was released in 1998 and it was published in the paper ''Microbial gene identification using interpolated Markov model''. Markov models were used to identify microbial genes in GLIMMER 1.0. GLIMMER considers the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mendelian Inheritance
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later popularized by William Bateson. These principles were initially controversial. When Mendel's theories were integrated with the Boveri–Sutton chromosome theory of inheritance by Thomas Hunt Morgan in 1915, they became the core of classical genetics. Ronald Fisher combined these ideas with the theory of natural selection in his 1930 book ''The Genetical Theory of Natural Selection'', putting evolution onto a mathematical footing and forming the basis for population genetics within the modern evolutionary synthesis. History The principles of Mendelian inheritance were named for and first derived by Gregor Johann Mendel, a nineteenth-century Moravian monk who formulated his ideas after conducting simple hybridisation experiments with pea plants (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
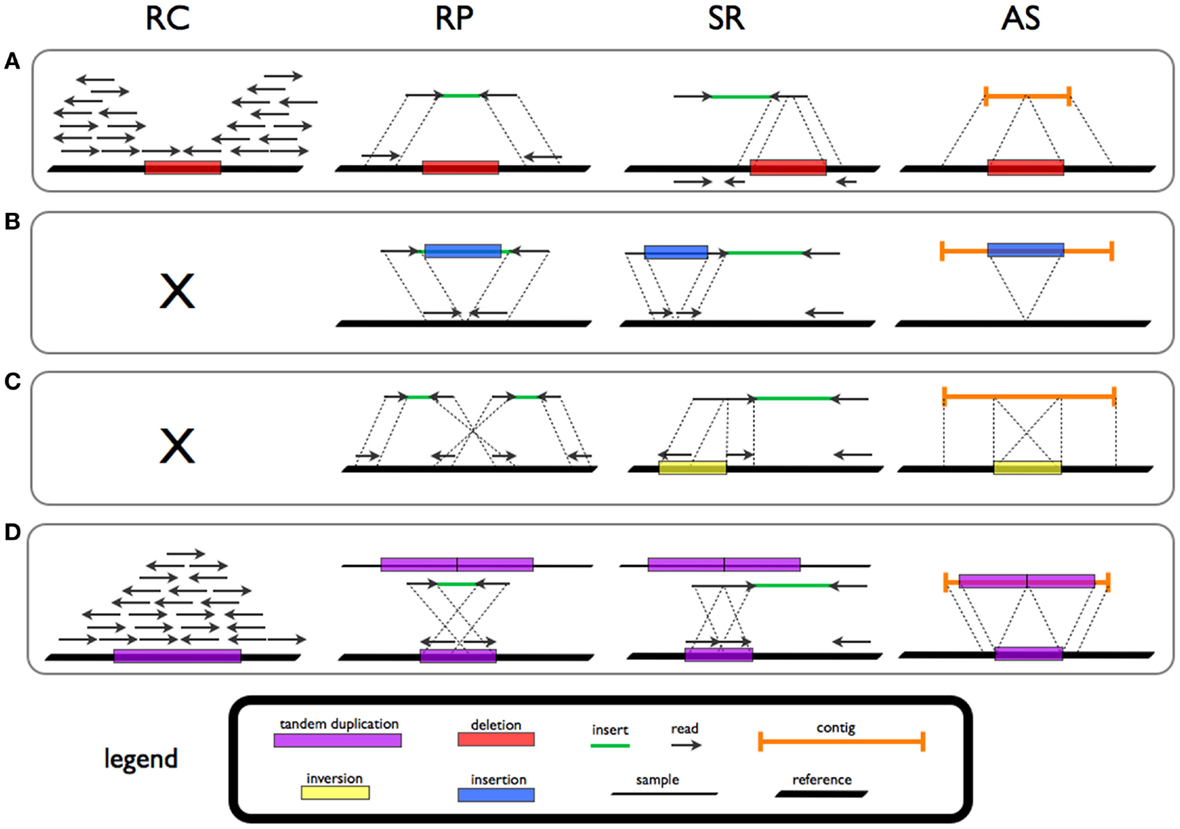
Structural Variations
Genomic structural variation is the variation in structure of an organism's chromosome. It consists of many kinds of variation in the genome of one species, and usually includes microscopic and submicroscopic types, such as deletions, duplications, copy-number variants, insertions, inversions and translocations. Originally, a structure variation affects a sequence length about 1kb to 3Mb, which is larger than SNPs and smaller than chromosome abnormality (though the definitions have some overlap). However, the operational range of structural variants has widened to include events > 50bp. The definition of structural variation does not imply anything about frequency or phenotypical effects. Many structural variants are associated with genetic diseases, however many are not. Recent research about SVs indicates that SVs are more difficult to detect than SNPs. Approximately 13% of the human genome is defined as structurally variant in the normal population, and there are at least 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was inv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
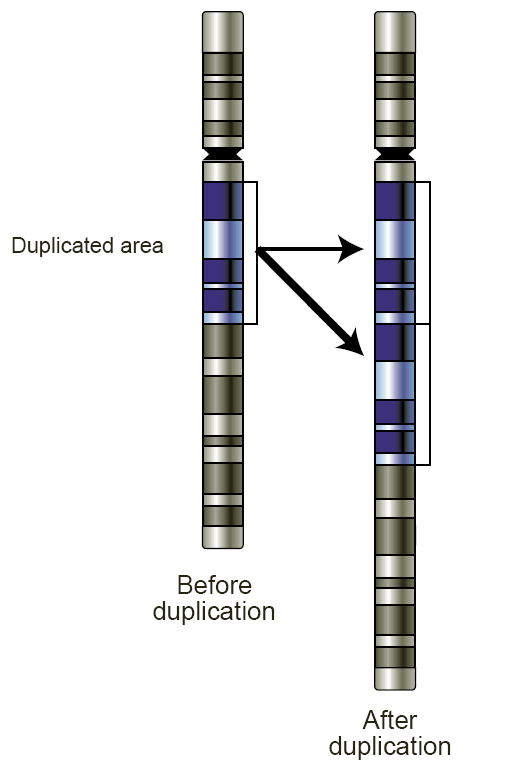
Copy-number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of Gene duplication, duplication or deletion (genetics), deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the locus (genetics), loci of interest. Short repeats include mainly Tandem repeat, dinucleotide repeats (two repeat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isochore (genetics)
In genetics, an isochore is a large region of genomic DNA (greater than 300 kilobases) with a high degree of uniformity in GC content; that is, guanine (G) and cytosine (C) bases. The distribution of bases within a genome is non-random: different regions of the genome have different amounts of G-C base pairs, such that regions can be classified and identified by the proportion of G-C base pairs they contain. Bernardi and colleagues first noticed the compositional non-uniformity of vertebrate genomes using thermal melting and density gradient centrifugation. The DNA fragments extracted by the gradient centrifugation were later termed "isochores", which was subsequently defined as "very long (much greater than 200 KB) DNA segments" that "are fairly homogeneous in base composition and belong to a small number of major classes distinguished by differences in guanine-cytosine (GC) content". Subsequently, the isochores "grew" and were claimed to be ">300 kb in size." The theory propos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |