|
Base Analog
Nucleic acid analogues are compounds which are Analog (chemistry), analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as peptide nucleic acid, PNA, which affect the properties of the chain (PNA can even form a triple helix). Nucleic acid analogues are also called Xeno Nucleic Acid and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries. Artificial nucleic acids include peptide nucleic ac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Difference DNA RNA-EN
Difference, The Difference, Differences or Differently may refer to: Music * ''Difference'' (album), by Dreamtale, 2005 * ''Differently'' (album), by Cassie Davis, 2009 ** "Differently" (song), by Cassie Davis, 2009 * ''The Difference'' (album), Pendleton, 2008 * "The Difference" (The Wallflowers song), 1997 * "The Difference", a song by Westlife from the 2009 album ''Where We Are'' * "The Difference", a song by Nick Jonas from the 2016 album ''Last Year Was Complicated'' * "The Difference", a song by Meek Mill featuring Quavo, from the 2016 mixtape '' DC4'' * "The Difference", a song by Matchbox Twenty from the 2002 album '' More Than You Think You Are'' * "The Difference", a 2020 song by Flume featuring Toro y Moi * "The Difference", a 2022 song by Ni/Co which represented Alabama in the ''American Song Contest'' * "Differences" (song), by Ginuwine, 2001 Science and mathematics * Difference (mathematics), the result of a subtraction * Difference equation, a type of recur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virus
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898,Dimmock p. 4 more than 9,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology. When infected, a host cell is often forced to rapidly produce thousands of copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles, or ''virions'', consisting of (i) the genetic material, i. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succinctly summarizes much of the atomic-level structure of the sequenced molecule. DNA sequencing DNA sequencing is the process of determining the nucleotide order of a given DNA fragment. So far, most DNA sequencing has been performed using the chain termination method developed by Frederick Sanger. This technique uses sequence-specific termination of a DNA synthesis reaction using modified nucleotide substrates. However, new sequencing technologies such as pyrosequencing are gaining an increasing share of the sequencing market. More genome data are now being produced by pyrosequencing than Sanger DNA sequencing. Pyrosequencing has enabled rapid genome sequencing. Bacterial genomes can be sequenced in a single run with several times cover ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dideoxynucleotides
Dideoxynucleotides are chain-elongating inhibitors of DNA polymerase, used in the Sanger method for DNA sequencing. They are also known as 2',3' because both the 2' and 3' positions on the ribose lack hydroxyl groups, and are abbreviated as ''ddNTPs'' (ddGTP, ddATP, ddTTP and ddCTP). Role in the Sanger method The Sanger method is used to amplify a target segment of DNA, so that the DNA sequence can be determined precisely. The incorporation of ddNTPs in the reaction valves are simply used to terminate the synthesis of a growing DNA strand, resulting in partially replicated DNA fragments. This is because DNA polymerase requires the 3' OH group of the growing chain and the 5' phosphate group of the incoming dNTP to create a phosphodiester bond. Sometimes the DNA polymerase will incorporate a ddNTP and the absence of the 3' OH group will interrupt the condensation reaction between the 5' phosphate (following the cleavage of pyrophospate) of the incoming nucleotide with the 3' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Synthesis
In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis (long peptides) in living organisms occurs in the opposite direction. The chemical synthesis of peptides can be carried out using classical solution-phase techniques, although these have been replaced in most research and development settings by solid-phase methods (see below). Solution-phase synthesis retains its usefulness in large-scale production of peptides for industrial purposes how ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide Synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides ( dA, dC, dG, and T), ribonucleosides ( A, C, G, and U), or chemically modified nucleosides, e.g. LNA or BNA. To obtain the desired oligonucleotide, the building blocks are sequentially coupled to the growing oligonucleotide chain in the order required by the sequence of the product (see ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Nucleic Acid
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA. Synthetic peptide nucleic acid oligomers have been used in recent years in molecular biology procedures, diagnostic assays, and antisense therapies. Due to their higher binding strength, it is not necessary to design long PNA oligomers for use in these roles, which usually require oligonucleotide probes of 20–25 bases. The main concern of the length of the PNA-oligomers is to guarantee the specificity. PNA oligomers also show greater specificity in binding to complementary DNAs, with a PNA/DNA base mismatch being more destabilizing than a similar mismatch in a DNA/DNA duplex. This binding strength and specificity also applies to PNA/RNA duplexes. PNAs are not easily recognized by either nucleases or proteases, making them resistant to degradation by enzymes. PNAs are also stable over a wide pH range. Though an unmodified PNA cannot readily cross the cell membrane to enter the cytosol, cov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Morpholino
A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule (colloquially, an oligo) used in molecular biology to modify gene expression. Its molecular structure contains DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups. Morpholinos block access of other molecules to small (~25 base) specific sequences of the base-pairing surfaces of ribonucleic acid (RNA). Morpholinos are used as research tools for reverse genetics by knocking down gene function. This article discusses only the Morpholino antisense oligomers, which are nucleic acid analogs. The word "Morpholino" can occur in other chemical names, referring to chemicals containing a six-membered morpholine ring. To help avoid confusion with other morpholine-containing molecules, when describing oligos "Morpholino" is often capitalized as a trade name, but this usage is not consistent across scienti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
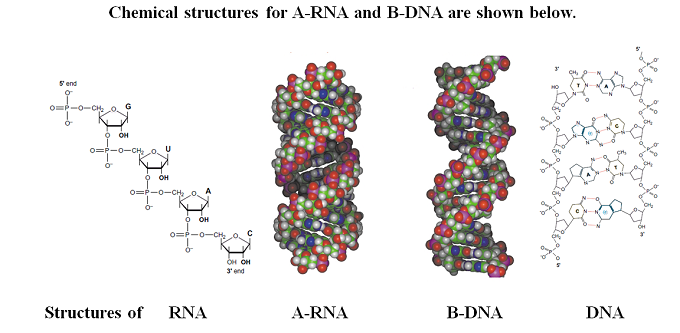
Bridged Nucleic Acid
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering. The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoamidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability. Chemical structures Chemical structures of BNA monomers containing a bridge at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer as synthesized by Takeshi Imanishi's group. The nature of the bridge can vary for different types of monomers. The 3D structures for A-RNA and B-DNA were used as a template for the design of the BNA monomers. The goal for the design was to find deriva ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalysis, catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate in a variety of RNA processing reactions, including RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligation (molecular Biology)
In molecular biology, ligation is the joining of two nucleic acid fragments through the action of an enzyme. It is an essential laboratory procedure in the molecular cloning of DNA, whereby DNA fragments are joined to create recombinant DNA molecules (such as when a foreign DNA fragment is inserted into a plasmid). The ends of DNA fragments are joined by the formation of phosphodiester bonds between the 3'-hydroxyl of one DNA terminus with the 5'-phosphoryl of another. RNA may also be ligated similarly. A co-factor is generally involved in the reaction, and this is usually ATP or NAD+. Ligation in the laboratory is normally performed using T4 DNA ligase. However, procedures for ligation without the use of standard DNA ligase are also popular. Ligation reaction The mechanism of the ligation reaction was first elucidated in the laboratory of I. Robert Lehman. Two fragments of DNA may be joined by DNA ligase which catalyzes the formation of a phosphodiester bond between the 3 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |