|
Biotin Carboxylase
In enzymology, a biotin carboxylase () is an enzyme that Catalysis, catalyzes the chemical reaction : ATP + biotin-carboxyl-carrier protein + CO2 \rightleftharpoons ADP + phosphate + carboxybiotin-carboxyl-carrier protein The three Substrate (biochemistry), substrates of this enzyme are Adenosine triphosphate, ATP, biotin-carboxyl-carrier protein (BCCP), and Carbon dioxide, CO2, whereas its three Product (chemistry), products are Adenosine diphosphate, ADP, phosphate, and carboxybiotin-carboxyl-carrier protein. The List of enzymes, systematic name of this enzyme class is biotin-carboxyl-carrier-protein:carbon-dioxide ligase (ADP-forming). This enzyme is also called biotin carboxylase (component of Acetyl-CoA carboxylase, acetyl CoA carboxylase). This enzyme participates in fatty acid biosynthesis. This enzyme participates in fatty acid biosynthesis by providing one of the catalytic functions of the Acetyl-CoA carboxylase complex. As previously mentioned, after the carboxybiot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphate
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid . The phosphate or orthophosphate ion is derived from phosphoric acid by the removal of three protons . Removal of one or two protons gives the dihydrogen phosphate ion and the hydrogen phosphate ion ion, respectively. These names are also used for salts of those anions, such as ammonium dihydrogen phosphate and trisodium phosphate. File:3-phosphoric-acid-3D-balls.png, Phosphoricacid File:2-dihydrogenphosphate-3D-balls.png, Dihydrogenphosphate File:1-hydrogenphosphate-3D-balls.png, Hydrogenphosphate File:0-phosphate-3D-balls.png, Phosphate In organic chemistry, phosphate or orthophosphate is an organophosphate, an ester of orthophosphoric acid of the form where one or more hydrogen atoms are replaced by organic groups. An example is trimethyl phosphate, . The term also refers to the triv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fatty Acid Biosynthesis
Fatty is a derogatory term for someone who is obese. It may refer also to: People * Mai Fatty, Gambian politician * Roscoe Arbuckle (1887–1933), American actor and comedian * Fatty Briody (1858–1903), American Major League Baseball player * Fatty D (April Fores), American pornographic actress * Bob Fothergill (1897–1938), American Major League Baseball outfielder * William Foulke (footballer) (1874–1916), English cricketer and footballer * Fatty George (1927–1982), Austrian jazz musician * Richard Lamb (1907–1974), Australian racing cyclist * Fatty Lawrence (1903–1976), college gridiron football player * W. T. McLain (1885–1938), college gridiron football player, lawyer, and politician * Charles H. Smith (American football), University of Michigan football player in 1893–1894 * Roland Taylor (1946–2017), retired American Basketball Association and National Basketball Association player * Paul Vautin (born 1959), Australian former rugby league footballer and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
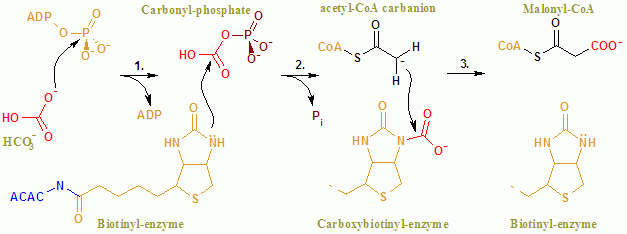
Acetyl-CoA Carboxylase
Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme () that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—'' ACACA'' and ''ACACB''. Structure Prokaryotes and plants have multi-subunit ACCs composed of several polypeptides. Biotin carboxylase (BC) activity, biotin carboxyl carrier protein (BCCP), and carboxyl transferase (CT) activity are each contained on a different s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Diphosphate
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenine attaches to the 1’ carbon. ADP can be interconverted to adenosine triphosphate (ATP) and adenosine monophosphate (AMP). ATP contains one more phosphate group than does ADP. AMP contains one fewer phosphate group. Energy transfer used by all living things is a result of dephosphorylation of ATP by enzymes known as ATPases. The cleavage of a phosphate group from ATP results in the coupling of energy to metabolic reactions and a by-product of ADP. ATP is continually reformed from lower-energy species ADP and AMP. The biosynthesis of ATP is achieved through ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |