|
Akkermansia Glycaniphila
''Akkermansia glycanphila'' is a species of intestinal mucin-degrading bacterium. It was first isolated from reticulated python ('' Malayopython reticulatus'') feces in 2016. Etymology The genus was named for Antoon DL Akkermans (1940–2006), a Dutch microbiologist recognized for his contribution to microbial ecology, and the epithet from the New Latin and Greek meaning "glycan-loving". Biology and biochemistry ''A. glycaniphila'', like, '' A. muciniphila'' is Gram-negative, strictly anaerobic, non-motile, non-spore-forming, oval-shaped bacterium. The typestrain is PytT (=DSM100705T=CIP 110913T). ''A. glycaniphila'' is able to use mucin as its sole source of carbon and nitrogen. It is culturable under the same conditions as ''A. muciniphilia'', (anaerobic conditions on medium containing gastric mucin). When grown on soft agar mucin medium, colonies appear white with a diameter of 0.7mm. The long axis of single cells is 0.6–1.0 μm. Cells are covered with filaments, and occur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coding Region
The coding region of a gene, also known as the coding sequence (CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the human genome and developing gene therapy. Definition Although this term is also sometimes used interchangeably with exon, it is not the exact same thing: the exon is composed of the coding region as well as the 3' and 5' untranslated regions of the RNA, and so therefore, an exon would be partially made up of coding regions. The 3' and 5' untranslated regions of the RNA, which do not code for protein, are termed non-coding regions and are not discussed on this page. There is often confusion between coding region ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxic
{{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aerobic Respiration
Cellular respiration is the process by which biological fuels are oxidised in the presence of an inorganic electron acceptor such as oxygen to produce large amounts of energy, to drive the bulk production of ATP. Cellular respiration may be described as a set of metabolic reactions and processes that take place in the cells of organisms to convert chemical energy from nutrients into adenosine triphosphate (ATP), and then release waste products. The reactions involved in respiration are catabolic reactions, which break large molecules into smaller ones, releasing energy. Respiration is one of the key ways a cell releases chemical energy to fuel cellular activity. The overall reaction occurs in a series of biochemical steps, some of which are redox reactions. Although cellular respiration is technically a combustion reaction, it is an unusual one because of the slow, controlled release of energy from the series of reactions. Nutrients that are commonly used by animal and pla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquinol Oxidase (H+-transporting)
A ubiquinol is an electron-rich (reduced) form of coenzyme Q (ubiquinone). The term most often refers to ubiquinol-10, with a 10-unit tail most commonly found in humans. The natural ubiquinol form of coenzyme Q is 2,3-dimethoxy-5-methyl-6-poly prenyl-1,4-benzoquinol, where the polyprenylated side-chain is 9-10 units long in mammals. Coenzyme Q10 (CoQ10) exists in three redox states, fully oxidized (ubiquinone), partially reduced (semiquinone or ubisemiquinone), and fully reduced (ubiquinol). The redox functions of ubiquinol in cellular energy production and antioxidant protection are based on the ability to exchange two electrons in a redox cycle between ubiquinol (reduced) and the ubiquinone (oxidized) form. Characteristics Because humans can synthesize ubiquinol, it is not classed as a vitamin. Bioavailability It is well-established that CoQ10 is not well absorbed into the body, as has been published in many peer-reviewed scientific journals. Since the ubiquinol form has tw ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
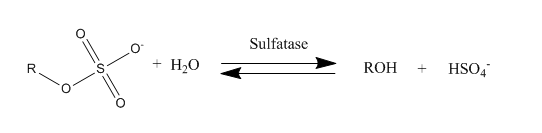
Sulfatases
Sulfatases are enzymes of the esterase class that catalyze the hydrolysis of sulfate esters. These may be found on a range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. In the latter case the resultant N-sulfates can also be termed sulfamates. Sulfatases play important roles in the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodelling sulfated glycosaminoglycans in the extracellular space. Together with sulfotransferases, sulfatases form the major catalytic machinery for the synthesis and breakage of sulfate esters. Occurrence and importance Sulfatases are found in lower and higher organisms. In higher organisms they are found in intracellular and extracellular spaces. Steroid sulfatase is distributed in a wide range of tissues throughout the body, enabling sulfated steroids synthesized in the adrenals and gonads t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neuraminidase
Exo-α-sialidase (EC 3.2.1.18, sialidase, neuraminidase; systematic name acetylneuraminyl hydrolase) is a glycoside hydrolase that cleaves the glycosidic linkages of neuraminic acids: : Hydrolysis of α-(2→3)-, α-(2→6)-, α-(2→8)- glycosidic linkages of terminal sialic acid residues in oligosaccharides, glycoproteins, glycolipids, colominic acid and synthetic substrates Neuraminidase enzymes are a large family, found in a range of organisms. The best-known neuraminidase is the viral neuraminidase, a drug target for the prevention of the spread of influenza infection. The viral neuraminidases are frequently used as antigenic determinants found on the surface of the influenza virus. Some variants of the influenza neuraminidase confer more virulence to the virus than others. Other homologues are found in mammalian cells, which have a range of functions. At least four mammalian sialidase homologues have been described in the human genome (see NEU1, NEU2, NEU3, NEU4). Sialidas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosyl Hydrolase
Glycoside hydrolases (also called glycosidases or glycosyl hydrolases) catalyze the hydrolysis of glycosidic bonds in complex sugars. They are extremely common enzymes with roles in nature including degradation of biomass such as cellulose (cellulase), hemicellulose, and starch (amylase), in anti-bacterial defense strategies (e.g., lysozyme), in pathogenesis mechanisms (e.g., viral neuraminidases) and in normal cellular function (e.g., trimming mannosidases involved in N-linked glycoprotein biosynthesis). Together with glycosyltransferases, glycosidases form the major catalytic machinery for the synthesis and breakage of glycosidic bonds. Occurrence and importance Glycoside hydrolases are found in essentially all domains of life. In prokaryotes, they are found both as intracellular and extracellular enzymes that are largely involved in nutrient acquisition. One of the important occurrences of glycoside hydrolases in bacteria is the enzyme beta-galactosidase (LacZ), wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycoside Hydrolases
Glycoside hydrolases (also called glycosidases or glycosyl hydrolases) catalyze the hydrolysis of glycosidic bonds in complex sugars. They are extremely common enzymes with roles in nature including degradation of biomass such as cellulose (cellulase), hemicellulose, and starch (amylase), in anti-bacterial defense strategies (e.g., lysozyme), in pathogenesis mechanisms (e.g., viral neuraminidases) and in normal cellular function (e.g., trimming mannosidases involved in N-linked glycoprotein biosynthesis). Together with glycosyltransferases, glycosidases form the major catalytic machinery for the synthesis and breakage of glycosidic bonds. Occurrence and importance Glycoside hydrolases are found in essentially all domains of life. In prokaryotes, they are found both as intracellular and extracellular enzymes that are largely involved in nutrient acquisition. One of the important occurrences of glycoside hydrolases in bacteria is the enzyme beta-galactosidase (LacZ), which i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operons
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but since the discovery of the first operons in eukaryotes in the early 1990s, more evidence has arisen to suggest they are more common than previously assumed. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNAs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create three-di ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |