|
ATAC-seq
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a technique used in molecular biology to assess genome-wide chromatin accessibility. In 2013, the technique was first described as an alternative advanced method for MNase-seq, FAIRE-Seq and DNase-Seq. ATAC-seq is a faster and more sensitive analysis of the epigenome than DNase-seq or MNase-seq. Description ATAC-seq identifies accessible DNA regions by probing open chromatin with hyperactive mutant Tn5 Transposase that inserts sequencing adapters into open regions of the genome. While naturally occurring transposases have a low level of activity, ATAC-seq employs the mutated hyperactive transposase. In a process called "tagmentation", Tn5 transposase cleaves and tags double-stranded DNA with sequencing adaptors. The tagged DNA fragments are then purified, PCR-amplified, and sequenced using next-generation sequencing. Sequencing reads can then be used to infer regions of increased accessibility as well as to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATAC-Seq Application V2
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a technique used in molecular biology to assess genome-wide chromatin accessibility. In 2013, the technique was first described as an alternative advanced method for MNase-seq, FAIRE-Seq and DNase-Seq. ATAC-seq is a faster and more sensitive analysis of the epigenome than DNase-seq or MNase-seq. Description ATAC-seq identifies accessible DNA regions by probing open chromatin with hyperactive mutant Tn5 Transposase that inserts sequencing adapters into open regions of the genome. While naturally occurring transposases have a low level of activity, ATAC-seq employs the mutated hyperactive transposase. In a process called "tagmentation", Tn5 transposase cleaves and tags double-stranded DNA with sequencing adaptors. The tagged DNA fragments are then purified, PCR-amplified, and sequenced using next-generation sequencing. Sequencing reads can then be used to infer regions of increased accessibility as well as to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MNase-seq
MNase-seq, short for micrococcal nuclease digestion with deep sequencing, is a molecular biological technique that was first pioneered in 2006 to measure nucleosome occupancy in the ''C. elegans'' genome, and was subsequently applied to the human genome in 2008. Though, the term ‘MNase-seq’ had not been coined until a year later, in 2009. Briefly, this technique relies on the use of the non-specific endo-exonuclease micrococcal nuclease, an enzyme derived from the bacteria ''Staphylococcus aureus'', to bind and cleave protein-unbound regions of DNA on chromatin. DNA bound to histones or other chromatin-bound proteins (e.g. transcription factors) may remain undigested. The uncut DNA is then purified from the proteins and sequenced through one or more of the various Next-Generation sequencing methods.'' MNase-seq is one of four classes of methods used for assessing the status of the epigenome through analysis of chromatin accessibility. The other three techniques are DNase-se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FAIRE-Seq
FAIRE-Seq (Formaldehyde-Assisted Isolation of Regulatory Elements) is a method in molecular biology used for determining the sequences of DNA regions in the genome associated with regulatory activity. The technique was developed in the laboratory of Jason D. Lieb at the University of North Carolina, Chapel Hill. In contrast to DNase-Seq, the FAIRE-Seq protocol doesn't require the permeabilization of cells or isolation of nuclei, and can analyse any cell type. In a study of seven diverse human cell types, DNase-seq and FAIRE-seq produced strong cross-validation, with each cell type having 1-2% of the human genome as open chromatin. Workflow The protocol is based on the fact that the formaldehyde cross-linking is more efficient in nucleosome-bound DNA than it is in nucleosome-depleted regions of the genome. This method then segregates the non cross-linked DNA that is usually found in open chromatin, which is then sequenced. The protocol consists of cross linking, phenol extraction a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transposition. The word "transposase" was first coined by the individuals who cloned the enzyme required for transposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that some segments of chromosomes changed their position, jumping between different loci or from one chromosome to another. The repositioning of these transposons (which coded for color) allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is als ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30-nm fiber. ''Current opinion in cell biology, 58,'' 95–104. https://doi.o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by Ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
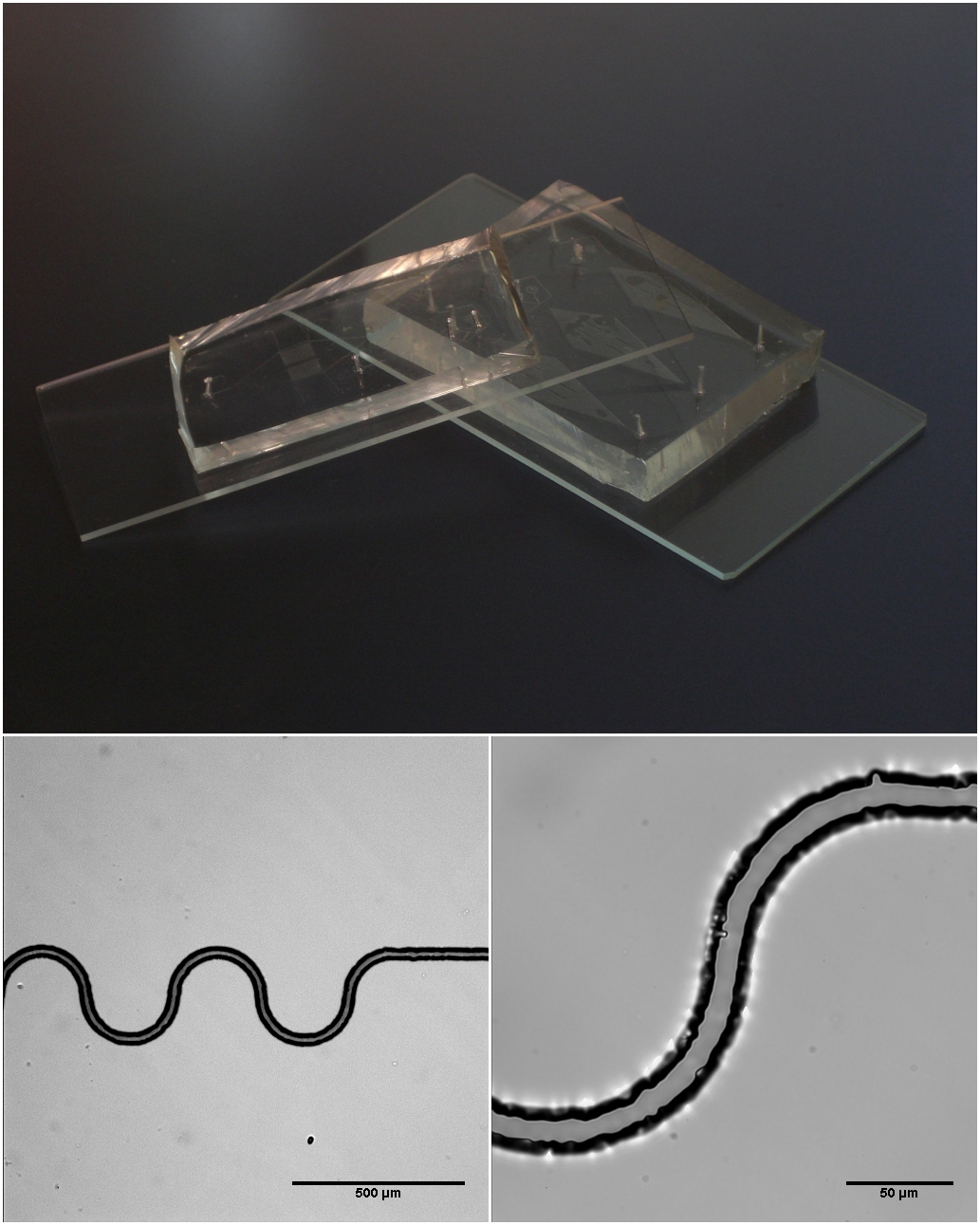
Microfluidics
Microfluidics refers to the behavior, precise control, and manipulation of fluids that are geometrically constrained to a small scale (typically sub-millimeter) at which surface forces dominate volumetric forces. It is a multidisciplinary field that involves engineering, physics, chemistry, biochemistry, nanotechnology, and biotechnology. It has practical applications in the design of systems that process low volumes of fluids to achieve multiplexing, automation, and high-throughput screening. Microfluidics emerged in the beginning of the 1980s and is used in the development of inkjet printheads, DNA chips, lab-on-a-chip technology, micro-propulsion, and micro-thermal technologies. Typically, micro means one of the following features: * Small volumes (μL, nL, pL, fL) * Small size * Low energy consumption * Microdomain effects Typically microfluidic systems transport, mix, separate, or otherwise process fluids. Various applications rely on passive fluid control using capillary fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-cell Analysis
In the field of cellular biology, single-cell analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is partic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macular Degeneration
Macular degeneration, also known as age-related macular degeneration (AMD or ARMD), is a medical condition which may result in blurred or no vision in the center of the visual field. Early on there are often no symptoms. Over time, however, some people experience a gradual worsening of vision that may affect one or both eyes. While it does not result in complete blindness, loss of central vision can make it hard to recognize faces, drive, read, or perform other activities of daily life. Visual hallucinations may also occur. Macular degeneration typically occurs in older people. Genetic factors and smoking also play a role. It is due to damage to the macula of the retina. Diagnosis is by a complete eye exam. The severity is divided into early, intermediate, and late types. The late type is additionally divided into "dry" and "wet" forms with the dry form making up 90% of cases. The difference between the two forms is the change of macula. Those with dry form AMD have drusen, ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis. As of 2016, two nucleobases have been found on which natural, enzymatic DNA methylation takes place: adenine and cytosine. The modified bases are N6-methyladenineD. B. Dunn, J. D. Smith: ''The occurrence of 6-methylaminopurine in deoxyribonucleic acids.'' In: ''Biochem J.'' 68(4), Apr 1958, S. 627–636. PMID 13522672. ., 5-methylcytosineB. F. Vanyushin, S. G. Tkacheva, A. N. Belozersky: ''Rare bases in animal DNA.'' In: ''Nature.'' 225, 1970, S. 948–949. PMID 4391887. and N4- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor-binding Site
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are typ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sonication
A sonicator at the Weizmann Institute of Science during sonicationSonication is the act of applying sound energy to agitate particles in a sample, for various purposes such as the extraction of multiple compounds from plants, microalgae and seaweeds. Ultrasonic frequencies (> 20 kHz) are usually used, leading to the process also being known as ultrasonication or ultra-sonication. In the laboratory, it is usually applied using an ''ultrasonic bath'' or an ''ultrasonic probe'', colloquially known as a ''sonicator''. In a paper machine, an ultrasonic foil can distribute cellulose fibres more uniformly and strengthen the paper. Effects Sonication has numerous effects, both chemical and physical. The chemical effects of ultrasound are concerned with understanding the effect of sonic waves on chemical systems, this is called sonochemistry. The chemical effects of ultrasound do not come from a direct interaction with molecular species. Studies have shown that no direct coupl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |