|
APOE (gene)
Apolipoprotein E (APOE) is a protein involved in the metabolism of fats in the body of mammals. A subtype is implicated in Alzheimer's disease and cardiovascular disease. APOE belongs to a family of fat-binding proteins called apolipoproteins. In the circulation, it is present as part of several classes of lipoprotein particles, including chylomicron remnants, VLDL, IDL, and some HDL. APOE interacts significantly with the low-density lipoprotein receptor (LDLR), which is essential for the normal processing (catabolism) of triglyceride-rich lipoproteins. In peripheral tissues, APOE is primarily produced by the liver and macrophages, and mediates cholesterol metabolism. In the central nervous system, APOE is mainly produced by astrocytes and transports cholesterol to neurons via APOE receptors, which are members of the low density lipoprotein receptor gene family. APOE is the principal cholesterol carrier in the brain. APOE is required for cholesterol transportation fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alzheimer's Disease
Alzheimer's disease (AD) is a neurodegeneration, neurodegenerative disease that usually starts slowly and progressively worsens. It is the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in short-term memory, remembering recent events. As the disease advances, symptoms can include primary progressive aphasia, problems with language, Orientation (mental), disorientation (including easily getting lost), mood swings, loss of motivation, self-neglect, and challenging behaviour, behavioral issues. As a person's condition declines, they often withdraw from family and society. Gradually, bodily functions are lost, ultimately leading to death. Although the speed of progression can vary, the typical life expectancy following diagnosis is three to nine years. The cause of Alzheimer's disease is poorly understood. There are many environmental and genetic risk factors associated with its development. The strongest genetic risk factor is from an alle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
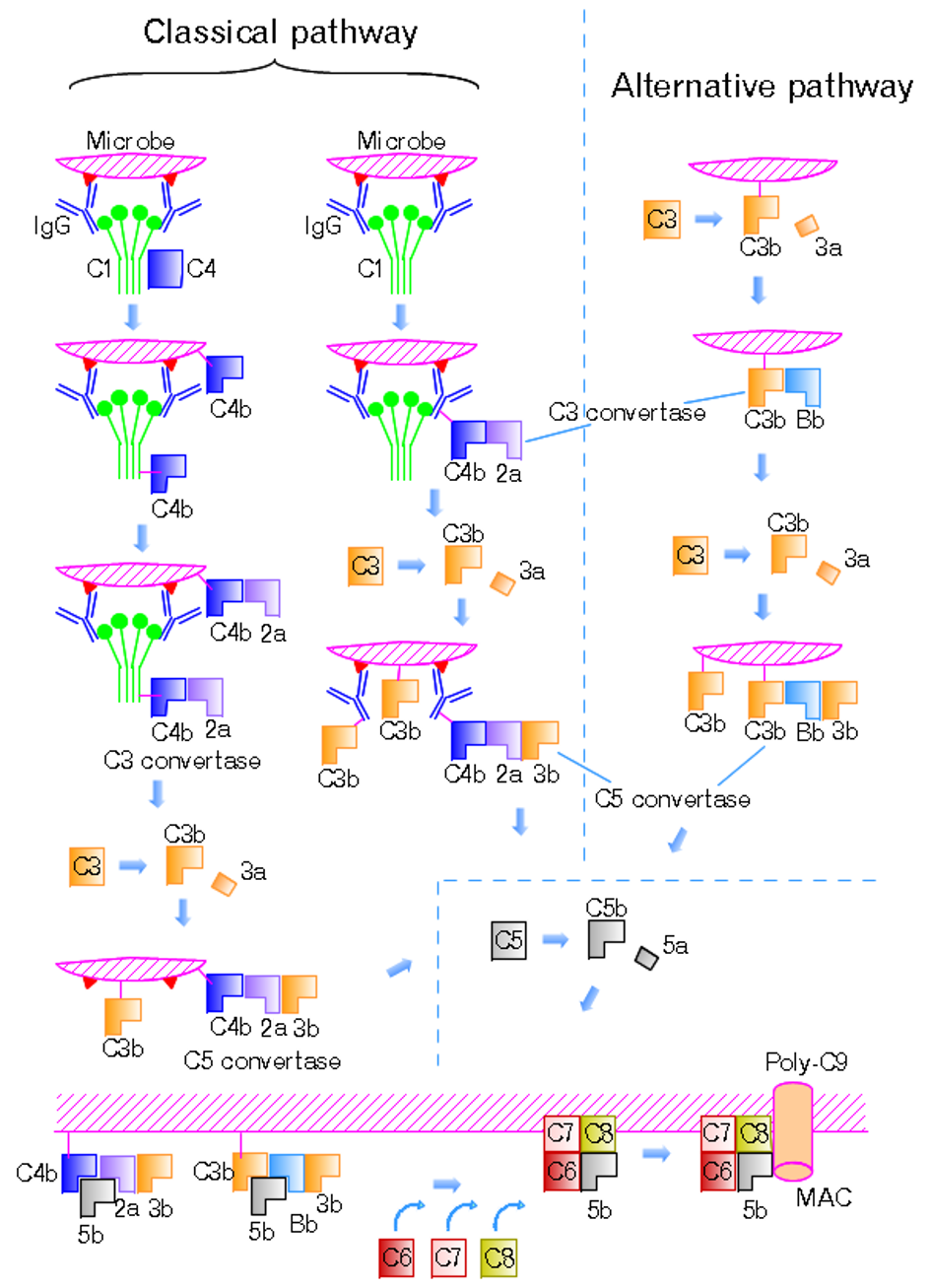
Classical Complement Pathway
The classical complement pathway is one of three pathways which activate the complement system, which is part of the immune system. The classical complement pathway is initiated by antigen-antibody complexes with the antibody isotypes IgG and IgM. Following activation, a series of proteins are recruited to generate C3 convertase (C4b2b, historically referred C4b2a), which cleaves the C3 protein. The C3b component of the cleaved C3 binds to C3 convertase (C4b2b) to generate C5 convertase (C4b2b3b), which cleaves the C5 protein. The cleaved products attract phagocytes to the site of infection and tags target cells for elimination by phagocytosis. In addition, the C5 convertase initiates the terminal phase of the complement system, leading to the assembly of the membrane attack complex ( MAC). The membrane attack complex creates a pore on the target cell's membrane, inducing cell lysis and death. The classical complement pathway can also be activated by apoptotic cells, necrotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Liver X Receptor
The liver X receptor (LXR) is a member of the nuclear receptor family of transcription factors and is closely related to nuclear receptors such as the PPARs, FXR and RXR. Liver X receptors (LXRs) are important regulators of cholesterol, fatty acid, and glucose homeostasis. LXRs were earlier classified as orphan nuclear receptors, however, upon discovery of endogenous oxysterols as ligands they were subsequently deorphanized. Two isoforms of LXR have been identified and are referred to as LXRα and LXRβ. The liver X receptors are classified into subfamily 1 (thyroid hormone receptor-like) of the nuclear receptor superfamily, and are given the nuclear receptor nomenclature symbols NR1H3 (LXRα) and NR1H2 (LXRβ) respectively. LXRα and LXRβ were discovered separately between 1994-1995. LXRα isoform was independently identified by two groups and initially named RLD-1 and LXR, whereas four groups identified the LXRβ isoform and called it UR, NER, OR-1, and RIP-15. The h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The Complementarity (molecular biology), complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Introns
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRNA introns, group I introns, group II introns, and s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apolipoprotein C2
Apolipoprotein C-II (Apo-CII, or Apoc-II), or apolipoprotein C2 is a protein that in humans is encoded by the gene. The protein encoded by this gene is secreted in plasma where it is a component of very low density lipoproteins and chylomicrons. This protein activates the enzyme lipoprotein lipase in capillaries, which hydrolyzes triglycerides and thus provides free fatty acids for cells. Mutations in this gene cause hyperlipoproteinemia type IB, characterized by xanthomas, pancreatitis, and hepatosplenomegaly, but no increased risk for atherosclerosis. Lab tests will show elevated blood levels of triglycerides, cholesterol, and chylomicrons Interactive pathway map See also * Apolipoprotein C In the field of molecular biology, apolipoprotein C is a family of four low molecular weight apolipoproteins, designated as C-I, C-II, C-III, and C-IV that are surface components of chylomicrons, VLDL, and HDL. In the fasting state, the C apol ... References * * * * * * * * * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apolipoprotein C1
Apolipoprotein C-I is a protein component of lipoproteins that in humans is encoded by the ''APOC1'' gene. Function The protein encoded by this gene is a member of the apolipoprotein C family. This gene is expressed primarily in the liver, and it is activated when monocytes differentiate into macrophages. Alternatively spliced transcript variants have been found for this gene, but the biological validity of some variants has not been determined. Apolipoprotein C-I has a length of 57 amino acids normally found in plasma and responsible for the activation of esterified lecithin cholesterol with an important role in the exchange of esterified cholesterol between lipoproteins and in removal of cholesterol from tissues. Its main function is inhibition of cholesteryl ester transfer protein (CETP), probably by altering the electric charge of HDL molecules. During fasting (like other apolipoprotein C), it is found primarily within HDL, while after a meal it is found on the surface ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Cluster
A gene family is a set of homologous genes within one organism. A gene cluster is a group of two or more genes found within an organism's DNA that encode similar polypeptides, or proteins, which collectively share a generalized function and are often located within a few thousand base pairs of each other. The size of gene clusters can vary significantly, from a few genes to several hundred genes. Portions of the DNA sequence of each gene within a gene cluster are found to be identical; however, the resulting protein of each gene is distinctive from the resulting protein of another gene within the cluster. Genes found in a gene cluster may be observed near one another on the same chromosome or on different, but homologous chromosomes. An example of a gene cluster is the Hox gene, which is made up of eight genes and is part of the Homeobox gene family. Formation Historically, four models have been proposed for the formation and persistence of gene clusters. Gene duplication and d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 19 (human)
Chromosome 19 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 19 spans more than 58.6 million base pairs, the building material of DNA. It is considered the most gene-rich chromosome containing roughly 1,500 genes, despite accounting for only 2 percent of the human genome. Genes Number of genes The following are some of the gene count estimates of human chromosome 19. Because researchers use different approaches to genome annotation, their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project ( CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes. Gene list The following is a partial list of genes on human chromosome 19. For complete list, see the link in the infobox on the right. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Choanoflagellate
The choanoflagellates are a group of free-living unicellular and colonial flagellate eukaryotes considered to be the closest living relatives of the animals. Choanoflagellates are collared flagellates, having a funnel shaped collar of interconnected microvilli at the base of a flagellum. Choanoflagellates are capable of both asexual and sexual reproduction. They have a distinctive cell morphology characterized by an ovoid or spherical cell body 3–10 µm in diameter with a single apical flagellum surrounded by a collar of 30–40 microvilli (see figure). Movement of the flagellum creates water currents that can propel free-swimming choanoflagellates through the water column and trap bacteria and detritus against the collar of microvilli, where these foodstuffs are engulfed. This feeding provides a critical link within the global carbon cycle, linking trophic levels. In addition to their critical ecological roles, choanoflagellates are of particular interest to evolutionary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |