|
APOBEC3
APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic subunit 3G) is a human enzyme encoded by the ''APOBEC3G'' gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity (medical), immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity. APOBEC3G exerts innate antiretroviral immune activity against retroviruses, most notably HIV, by interfering with proper replication. However, lentiviruses such as HIV have evolved the Viral infectivity factor (Vif) protein in order to counteract this effect. Vif interacts with APOBEC3G and triggers the ubiquitination and degradation of APOBEC3G via the proteasomal pathway. On the other hand, Human foamy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
APOBEC
image:Apobec.J.Steinfeld.D.png, 300px, upExample of a member of the APOBEC family, APOBEC-2. A cytidine deaminase from ''Homo sapiens''.; ; rendered usinPyMOL APOBEC ("apolipoprotein B mRNA editing enzyme, catalytic polypeptide") is a family of evolutionarily conserved cytidine deaminases. A mechanism of generating protein diversity is messenger RNA, mRNA editing. Members of this family are RNA editing#C-U editing, C-to-U editing enzymes. The N-terminal domain of APOBEC like proteins is the catalytic domain, while the C-terminal domain is a pseudocatalytic domain. More specifically, the catalytic domain is a zinc dependent cytidine deaminase domain and is essential for cytidine deamination. RNA editing by APOBEC-1 requires homodimerisation and this complex interacts with RNA binding proteins to form the editosome. In humans/mammals they help protect from viral infections. These enzymes, when misregulated, are a major source of mutation in numerous cancer types. A 2013 revi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Infectivity Factor
Viral infectivity factor, or Vif, is an accessory protein found in HIV and other lentiviruses. Its role is to disrupt the antiviral activity of the human enzyme APOBEC (specifically APOBEC3G, "A3G" in short) by targeting it for ubiquitination and cellular degradation. APOBEC is a cytidine deaminase enzyme that mutates viral nucleic acids. Vif is a 23-kilodalton protein that is essential for viral replication. Vif inhibits the cellular protein, APOBEC3G, from entering the virion during budding from a host cell by targeting it for proteasomal degradation. Vif binds to A3G as well as the cellular Cullin5 E3 Ubiquitin Ligase ( ELOB- ELOC-CUL5) and a CBFB cofactor so that the ligase can be hijacked to tag A3G for degradation. The crystal Structure of the HIV Vif BC-box in Complex with Human Elongin B and Elongin C was solved in 2008, and the structure of the full Vif/E3 complex was solved in 2014. In the absence of Vif, APOBEC3G causes hypermutation of the viral genome, rendering it d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Foamy Virus
Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus ''Spumavirus''. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection. Discovery The first description of foamy virus (FV) was in 1954. It was found as a contaminant in primary monkey kidney cultures. The first isolate of the “foamy viral agent” was in 1955. Not too long after this, it was isolated from a wide variety of New and Old World monkeys, cats, and cows. It was not until several years later that humans entered the scene. In 1971, a viral agent with FV-like characteristics was isolated from lymphoblastoid cells released from a human nasopharyngeal carcinoma (NPC) from a Kenyan patient. The agent was termed a human FV bec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lentiviruses
''Lentivirus'' is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals. Lentiviruses can integrate a significant amount of viral complementary DNA into the DNA of the host cell and can efficiently infect nondividing cells, so they are one of the most efficient methods of gene delivery. They can become endogenous, integrating their genome into the host germline genome, so that the virus is henceforth inherited by the host's descendants. Classification Five serogroups of lentiviruses are recognized, reflecting the vertebrate hosts with which they are associated (primates, sheep and goats, horses, domestic cats, and cattle). The primate lentiviruses are distinguished ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
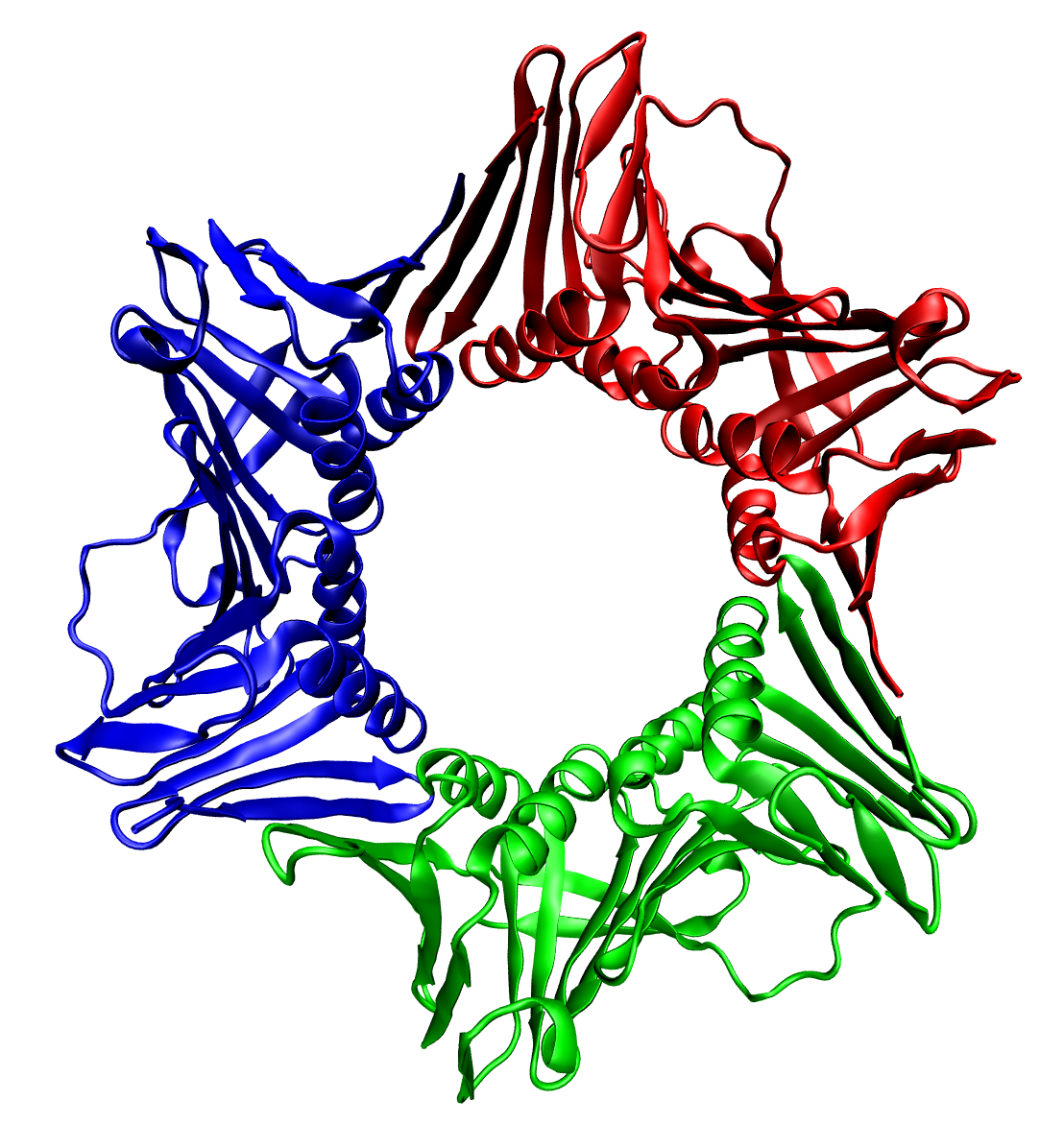
Protein Trimer
In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein. Porins usually arrange themselves in membranes as trimers. Bacteriophage T4 tail fiber Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation. The distal portion of each of the bacteriophage T4 tail fibers is encoded by ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tetramer
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin). Subunit interactions in tetramers The interactions between subunits forming a tetramer is primarily determined by non covalent interaction. Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as Sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Encapsidation
{{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |