|
A2 Holin Family
The Putative Archaeal 2 TMS Holin (A2-Hol) FamilyTC# 9.B.109 consists of a few putative holins from '' Nitrososphaerota'' ranging in size from about 130 to 165 amino acyl residues (aas) and exhibiting 2 transmembrane segments (TMSs). A representative list of proteins belonging to the A2-Hol family can be found in thTransporter Classification Database The archaeon, Candidatus ''Nitrosoarchaeum limnia'', encodes adjacent genes designated Toxin Secretion/Lysis Holin. The "toxin" gene encodes a soluble protein of 325 aas stated as belonging to the "Glycosyltransferase GBT-type Superfamily". This protein brings up other glycosyltransferases in a NCBI BLAST search. The adjacent gene encodes a small protein of 132 aas and 2 TMSs TC# 9.B.109.1.1 that could be a holin, based on its size and topology. This protein has the UniProt accession number oS2E3C4 Paralogues are found in this same organism (Candidatus ''Nitrosoarchaeum koreensis'') and other closely related species. See also * Ho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitrososphaerota
The Nitrososphaerota (syn. Thaumarchaeota) are a phylum of the Archaea proposed in 2008 after the genome of ''Cenarchaeum symbiosum'' was sequenced and found to differ significantly from other members of the hyperthermophilic phylum Thermoproteota (formerly Crenarchaeota). Three described species in addition to ''C. symbiosum'' are ''Nitrosopumilus maritimus'', ''Nitrososphaera viennensis'', and ''Nitrososphaera gargensis''. The phylum was proposed in 2008 based on phylogenetic data, such as the sequences of these organisms' ribosomal RNA genes, and the presence of a form of type I topoisomerase that was previously thought to be unique to the eukaryotes. This assignment was confirmed by further analysis published in 2010 that examined the genomes of the ammonia-oxidizing archaea ''Nitrosopumilus maritimus'' and ''Nitrososphaera gargensis'', concluding that these species form a distinct lineage that includes ''Cenarchaeum symbiosum''. The lipid crenarchaeol has been found only i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of ''Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosyltransferase
Glycosyltransferases (GTFs, Gtfs) are enzymes ( EC 2.4) that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar (also known as the "glycosyl donor") to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based. The result of glycosyl transfer can be a carbohydrate, glycoside, oligosaccharide, or a polysaccharide. Some glycosyltransferases catalyse transfer to inorganic phosphate or water. Glycosyl transfer can also occur to protein residues, usually to tyrosine, serine, or threonine to give O-linked glycoproteins, or to asparagine to give N-linked glycoproteins. Mannosyl groups may be transferred to tryptophan to generate C-mannosyl tryptophan, which is relatively abundant in eukaryotes. Transferases may also use lipids as an acceptor, forming glycolipids, and even use lipid-linked sugar phosphate donors, such as dolichol phosphates in eukaryotic o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matching p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Holin
Holins are a diverse group of small proteins produced by dsDNA bacteriophages in order to trigger and control the degradation of the host's cell wall at the end of the lytic cycle. Holins form pores in the host's cell membrane, allowing lysins to reach and degrade peptidoglycan, a component of bacterial cell walls. Holins have been shown to regulate the timing of lysis with great precision. Over 50 unrelated gene families encode holins, making them the most diverse group of proteins with common function. Together with lysins, holins are being studied for their potential use as antibacterial agents. While canonical holins act by forming large pores, pinholins such as the S protein of lambdoid phage 21 act by forming heptameric channels that depolarize the bacterial membrane. They are associated with SAR endolysins, which remain inactive in the periplasm prior to the depolarization of the membrane. Viruses that infect eukaryotic cells may use similar channel-forming proteins called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysin
Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins. Lysins are being used as antibacterial agents due to their high effectiveness and specificity in comparison with antibiotics, which are susceptible to bacterial resistance. Structure Double-stranded DNA phage lysins tend to lie within the 25 to 40 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein familie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membrane Proteins
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the surv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Proteins
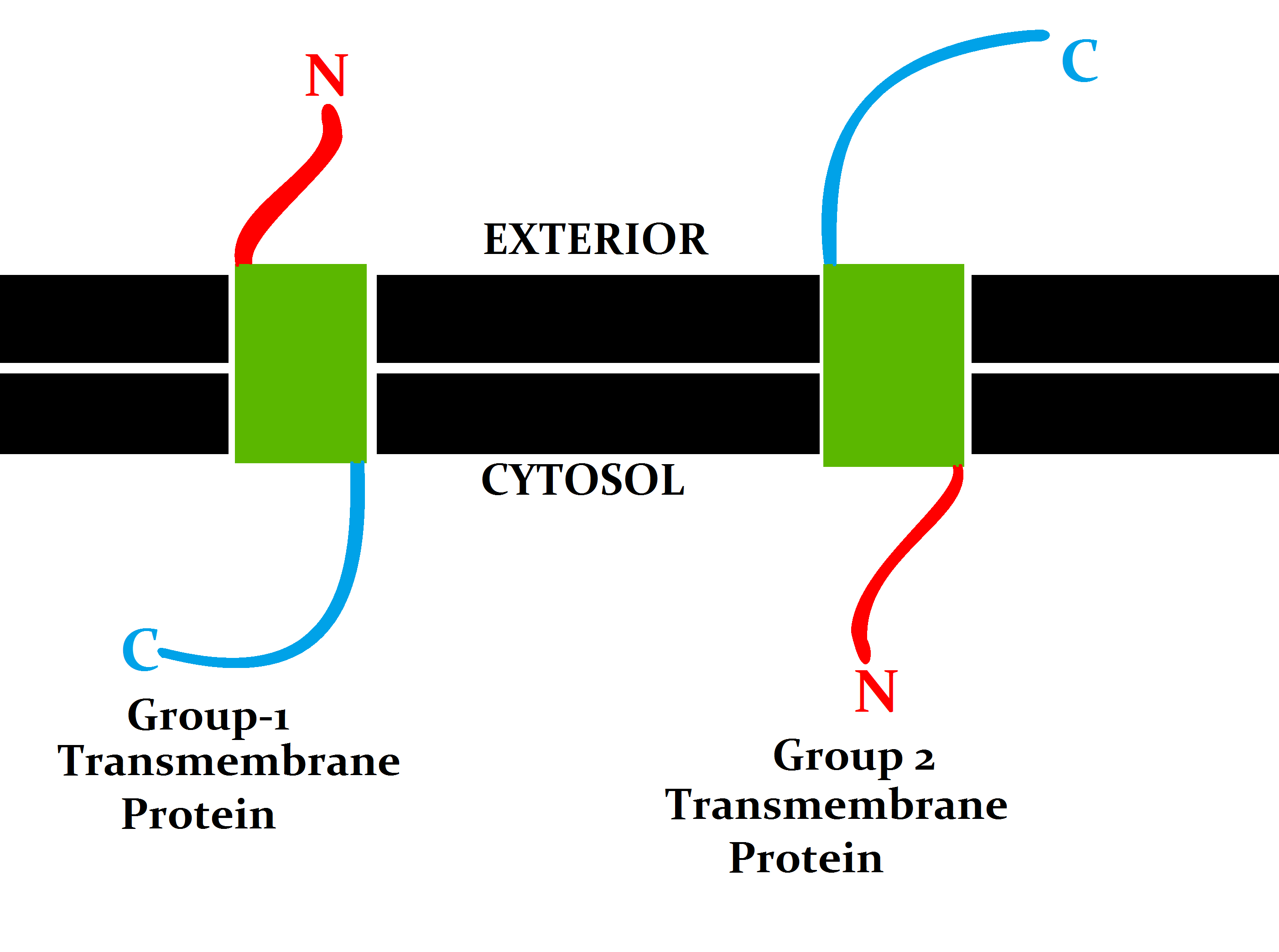
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Transporters
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transport Proteins
A transport protein (variously referred to as a transmembrane pump, transporter, escort protein, acid transport protein, cation transport protein, or anion transport protein) is a protein that serves the function of moving other materials within an organism. Transport proteins are vital to the growth and life of all living things. There are several different kinds of transport proteins. Carrier proteins are proteins involved in the movement of ions, small molecules, or macromolecules, such as another protein, across a biological membrane. Carrier proteins are integral membrane proteins; that is, they exist within and span the membrane across which they transport substances. The proteins may assist in the movement of substances by facilitated diffusion (i.e., passive transport) or active transport. These mechanisms of movement are known as carrier-mediated transport. Each carrier protein is designed to recognize only one substance or one group of very similar substances. Research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)