|
Astroviridae Infections
Astroviruses are a type of virus that was first discovered in 1975 using electron microscopes following an outbreak of diarrhea in humans. In addition to humans, astroviruses have now been isolated from numerous mammalian animal species (and are classified as genus '' Mamastrovirus'') and from avian species such as ducks, chickens, and turkey poults (classified as genus ''Avastrovirus''). Astroviruses are 28–35 nm diameter, icosahedral viruses that have a characteristic five- or six-pointed star-like surface structure when viewed by electron microscopy. Along with the Picornaviridae and the Caliciviridae, the Astroviridae comprise a third family of nonenveloped viruses whose genome is composed of plus- sense, single-stranded RNA. Astrovirus has a non-segmented, single stranded, positive sense RNA genome within a non-enveloped icosahedral capsid. Human astroviruses have been shown in numerous studies to be an important cause of gastroenteritis in young children worldwi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmission Electron Microscopy
Transmission electron microscopy (TEM) is a microscopy technique in which a beam of electrons is transmitted through a specimen to form an image. The specimen is most often an ultrathin section less than 100 nm thick or a suspension on a grid. An image is formed from the interaction of the electrons with the sample as the beam is transmitted through the specimen. The image is then magnified and focused onto an imaging device, such as a fluorescent screen, a layer of photographic film, or a sensor such as a scintillator attached to a charge-coupled device. Transmission electron microscopes are capable of imaging at a significantly higher resolution than light microscopes, owing to the smaller de Broglie wavelength of electrons. This enables the instrument to capture fine detail—even as small as a single column of atoms, which is thousands of times smaller than a resolvable object seen in a light microscope. Transmission electron microscopy is a major analytical method i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fecal–oral Route
The fecal–oral route (also called the oral–fecal route or orofecal route) describes a particular route of transmission of a disease wherein pathogens in fecal particles pass from one person to the mouth of another person. Main causes of fecal–oral disease transmission include lack of adequate sanitation (leading to open defecation), and poor hygiene practices. If soil or water bodies are polluted with fecal material, humans can be infected with waterborne diseases or soil-transmitted diseases. Fecal contamination of food is another form of fecal-oral transmission. Washing hands properly after changing a baby's diaper or after performing anal hygiene can prevent foodborne illness from spreading. The common factors in the fecal-oral route can be summarized as five Fs: fingers, flies, fields, fluids, and food. Diseases caused by fecal-oral transmission include typhoid, cholera, polio, hepatitis and many other infections, especially ones that cause diarrhea. Background Al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potyviridae
''Potyviridae'' is a family of positive-strand RNA viruses that encompasses more than 30% of known plant viruses, many of which are of great agricultural significance. The family has 12 genera and 235 species, three of which are unassigned to a genus. Structure Potyvirid virions are nonenveloped, flexuous filamentous, rod-shaped particles. The diameter is around 12–15 nm, with a length of 200–300 nm. Genome Genomes are linear and usually nonsegmented, around 8–12kb in length, consisting of positive-sense RNA, which is surrounded by a protein coat made up of a single viral encoded protein called a capsid. All induce the formation of virus inclusion bodies called cylindrical inclusions (‘pinwheels’) in their hosts. These consist of a single protein (about 70 kDa) made in their hosts from a single viral genome product. Member viruses encode large polypeptides that are cleaved into mature proteins. In 5'–3' order these proteins are * P1 (a serine protease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tymoviridae
''Tymoviridae'' is a family of single-stranded positive sense RNA viruses in the order ''Tymovirales''. Plants serve as natural hosts. There are 42 species in this family, assigned to three genera, with two species unassigned to a genus. Taxonomy The family includes the following three genera: * ''Maculavirus'' * ''Marafivirus'' * ''Tymovirus'' Additionally, the following two species are not assigned to a genus: * ''Bombyx mori latent virus'' * ''Poinsettia mosaic virus'' Proposed viruses * Culex tymovirusWang L, Lv X, Zhai Y, Fu S, Wang D, Rayner S, Tang Q, Liang G (2012) Genomic characterization of a novel virus of the family tymoviridae isolated from mosquitoes. PLoS One 7(7):e39845. * Fig fleck-associated virus Virology The virions are non-enveloped and isometry, isometric with a diameter of around 30 nm, with an icosahedral structure and a Capsid#Triangulation number, triangulation number T=3. The linear genome is between of 6–7.5 kilobases in length and encodes o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) ''interchromosomal'' recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes (random orientation of pairs of homologous chromosomes in meiosis I); & (2) ''intrachromosomal'' recombination, occurring through crossing over. During meiosis in eukaryotes, genetic recombination involves the pairing of homologous chromosomes. This may be followed by information transfer between the chromosomes. The information transfer may occur without physical exchange (a se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
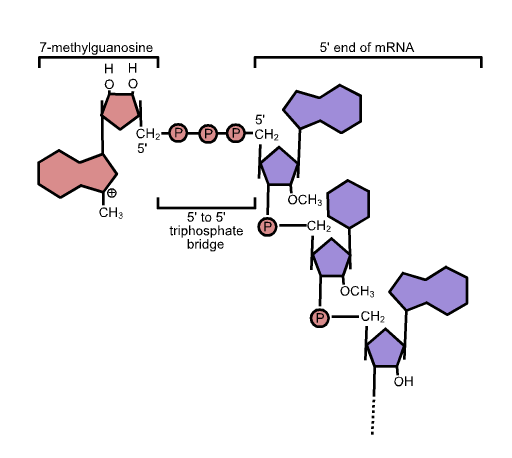
5' Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2′-hydroxy group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3' End
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poly A
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nuclear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-dependent RNA Polymerase
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template. RdRp is an essential protein encoded in the genomes of most RNA-containing viruses with no DNA stage including SARS-CoV-2. Some eukaryotes also contain RdRps, which are involved in RNA interference and differ structurally from viral RdRps. History Viral RdRps were discovered in the early 1960s from studies on mengovirus and polio virus when it was observed that these viruses were not sensitive to actinomycin D, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested that there is a virus-specific enzyme that could copy RNA from an RNA template and not from a DNA template. Distr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |