|
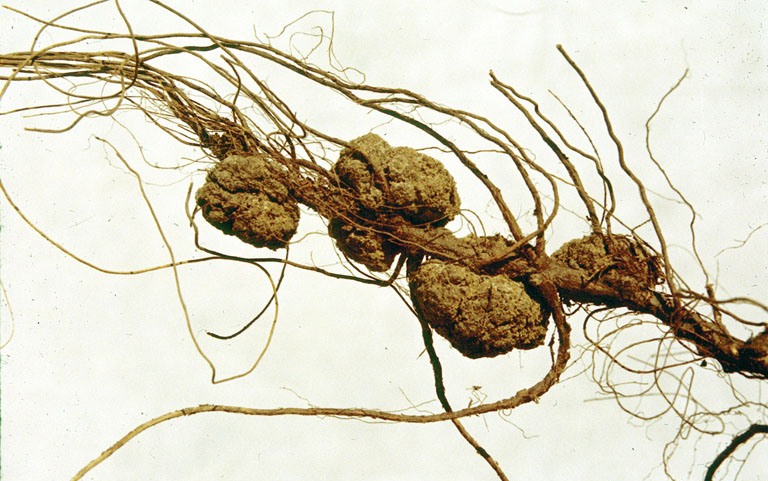
Agrobacterium Skierniewicense
''Agrobacterium'' is a genus of Gram-negative bacteria established by H. J. Conn that uses horizontal gene transfer to cause tumors in plants. ''Agrobacterium tumefaciens'' is the most commonly studied species in this genus. ''Agrobacterium'' is well known for its ability to transfer DNA between itself and plants, and for this reason it has become an important tool for genetic engineering. Nomenclatural History Leading up to the 1990s, the genus ''Agrobacterium'' was used as a wastebasket taxon. With the advent of 16S sequencing, many ''Agrobacterium'' species (especially the marine species) were reassigned to genera such as ''Ahrensia'', ''Pseudorhodobacter'', ''Ruegeria'', and ''Stappia''. The remaining ''Agrobacterium'' species were assigned to three biovars: biovar 1 (''Agrobacterium tumefaciens''), biovar 2 (''Agrobacterium rhizogenes''), and biovar 3 (''Agrobacterium vitis''). In the early 2000s, ''Agrobacterium'' was synonymized with the genus ''Rhizobium''. This move pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Agrobacterium Radiobacter
''Agrobacterium radiobacter'' (more commonly known as ''Agrobacterium tumefaciens'') is the causal agent of crown gall disease (the formation of tumours) in over 140 species of eudicots. It is a rod-shaped, Gram-negative soil bacterium. Symptoms are caused by the insertion of a small segment of DNA (known as the T-DNA, for 'transfer DNA', not to be confused with tRNA that transfers amino acids during protein synthesis), from a plasmid into the plant cell, which is incorporated at a semi-random location into the plant genome. Plant genomes can be engineered by use of ''Agrobacterium'' for the delivery of sequences hosted in T-DNA binary vectors. ''Agrobacterium tumefaciens'' is an Alphaproteobacterium of the family Rhizobiaceae, which includes the nitrogen-fixing legume symbionts. Unlike the nitrogen-fixing symbionts, tumor-producing ''Agrobacterium'' species are pathogenic and do not benefit the plant. The wide variety of plants affected by ''Agrobacterium'' makes it of gr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Horizontal Gene Transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between Unicellular organism, unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher order evolution while more significantly shifting perspectives on bacterial evolution. Horizontal gene transfer is the primary mechanism for the spread of antibiotic resistance in bacteria, and plays an important role in the evolution of bacteria that can degrade novel compounds such as human-created Bactericide, pesticides and in the evolution, maintenance, and transmission of virulence. It often involves Temperateness (virology), temperate bacteriophages and plasmids. Genes responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synapomorphy
In phylogenetics, an apomorphy (or derived trait) is a novel character or character state that has evolved from its ancestral form (or plesiomorphy). A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor. ) In cladistics, synapomorphy implies homology. Examples of apomorphy are the presence of erect gait, fur, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a sprawling gait and lack of fur. Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals. Etymology The word —coined by German entomologist Willi Hennig—is derived from the Ancient Greek words (''sún''), meaning "with, together"; (''apó''), meaning "away from"; and (''morphḗ''), meaning "shape, form". Clade analysis T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monophyly
In cladistics for a group of organisms, monophyly is the condition of being a clade—that is, a group of taxa composed only of a common ancestor (or more precisely an ancestral population) and all of its lineal descendants. Monophyletic groups are typically characterised by shared derived characteristics ( synapomorphies), which distinguish organisms in the clade from other organisms. An equivalent term is holophyly. The word "mono-phyly" means "one-tribe" in Greek. Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic group'' consists of all of the descendants of a common ancestor minus one or more monophyletic groups. A '' polyphyletic group'' is characterized by convergent features or habits of scientific interest (for example, night-active primates, fruit trees, aquatic insects). The features by which a polyphyletic group is differentiated from others are not inherited from a common ancestor. These definitions have taken ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhizobium
''Rhizobium'' is a genus of Gram-negative soil bacteria that fix nitrogen. ''Rhizobium'' species form an endosymbiotic nitrogen-fixing association with roots of (primarily) legumes and other flowering plants. The bacteria colonize plant cells within root nodules, where they convert atmospheric nitrogen into ammonia using the enzyme nitrogenase and then provide organic nitrogenous compounds such as glutamine or ureides to the plant. The plant, in turn, provides the bacteria with organic compounds made by photosynthesis. This mutually beneficial relationship is true of all of the rhizobia, of which the genus ''Rhizobium'' is a typical example. ''Rhizobium'' is also capable to solubilize phosphorus. History Martinus Beijerinck was the first to isolate and cultivate a microorganism from the nodules of legumes in 1888. He named it ''Bacillus radicicola'', which is now placed in ''Bergey's Manual of Determinative Bacteriology'' under the genus ''Rhizobium''. Research ''Rhizobium'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stappia
In taxonomy, ''Stappia'' is a genus of the Hyphomicrobiales The ''Hyphomicrobiales'' are an order of Gram-negative Alphaproteobacteria. The rhizobia, which fix nitrogen and are symbiotic with plant roots, appear in several different families. The four families ''Nitrobacteraceae'', ''Hyphomicrobiaceae' .... Some members of the genus (now transferred to '' Labrenzia'') oxidize carbon monoxide (CO) aerobically. Stappia indica is a diatom associated bacterium which is known to inhibit the growth of diatoms such as Thalassiosira pseudonana. References Further reading Scientific journals * * Scientific books * Scientific databases External links Bacteria genera Hyphomicrobiales {{Hyphomicrobiales-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ruegeria
In taxonomy, ''Ruegeria'' is a genus of the Rhodobacteraceae. This genus was formerly known as the marine ''Agrobacterium'' before they were reclassified in 1998. It bears in fact the name of Hans-Jürgen Rüger, a German microbiologist, for his contribution to the taxonomy of marine species of ''Agrobacterium''. Characteristics The genus is characterised by members who are: * Gram-negative (like all Proteobacteria) * ovoid to rod-shaped cells 0.6–1.6 × 1.0–4.0 µm * motile by polar flagella, or nonmotile * non-sporeformers * aerobic * oxidase and catalase positive * are incapable of photosynthetic growth * bacteriochlorophyll a is absent * major quinone is ubiquinone 10 * mol% G+C of the DNA is 55–59 * member of the family ''Rhodobacteraceae'', which is phenotypically, metabolically, ecologically diverse and defined based on 16S sequences data Phylogenetically it is very close to the genus ''Silicibacter'', but the two remain separate species due to phenotypic dif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudorhodobacter
In taxonomy, ''Pseudorhodobacter'' is a genus of the Rhodobacteraceae.See the NCBI The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The ...br>webpage on Pseudorhodobacter Data extracted from the References Further reading Scientific journals * Scientific books * Scientific databases External links Rhodobacteraceae Bacteria genera {{Rhodobacterales-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ahrensia
In taxonomy, ''Ahrensia'' is a genus of the Hyphomicrobiales.See the NCBIbr>webpage on Ahrensia Data extracted from the ''Ahrensia'' is named after the German microbiologist R. Ahrens. The cells are rod-shaped and motile. They are strictly aerobic.Garrity, George M.; Brenner, Don J.; Krieg, Noel R.; Staley, James T. (eds.) (2005). Bergey's Manual of Systematic Bacteriology, Volume Two: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. New York, New York: Springer. . See also * List of bacterial genera named after personal names Many bacterial species are named after people, either the discoverer or a famous person in the field of microbiology. For example, ''Salmonella'' is named after D.E. Salmon, who discovered it (albeit as "''Bacillus typhi''"). For the generic epit ... References Further reading * * External links Hyphomicrobiales Bacteria genera {{Hyphomicrobiales-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wastebasket Taxon
Wastebasket taxon (also called a wastebin taxon, dustbin taxon or catch-all taxon) is a term used by some taxonomists to refer to a taxon that has the sole purpose of classifying organisms that do not fit anywhere else. They are typically defined by either their designated members' often superficial similarity to each other, or their ''lack'' of one or more distinct character states or by their ''not'' belonging to one or more other taxa. Wastebasket taxa are by definition either paraphyletic or polyphyletic, and are therefore not considered valid taxa under strict cladistic rules of taxonomy. The name of a wastebasket taxon may in some cases be retained as the designation of an evolutionary grade, however. The term was coined in a 1985 essay by Steven Jay Gould. Examples There are many examples of paraphyletic groups, but true "wastebasket" taxa are those that are known not to, and perhaps not intended to, represent natural groups, but are nevertheless used as convenient groups ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including the transfer of genes within and across species boundaries to produce improved or novel organisms. New DNA is obtained by either isolating and copying the genetic material of interest using recombinant DNA methods or by artificially synthesising the DNA. A construct is usually created and used to insert this DNA into the host organism. The first recombinant DNA molecule was made by Paul Berg in 1972 by combining DNA from the monkey virus SV40 with the lambda virus. As well as inserting genes, the process can be used to remove, or "knock out", genes. The new DNA can be inserted randomly, or targeted to a specific part of the genome. An organism that is generated through genetic engineering is considered to be genetically modified (GM) an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |