|
Accessible Surface Area
The accessible surface area (ASA) or solvent-accessible surface area (SASA) is the surface area of a biomolecule that is accessible to a solvent. Measurement of ASA is usually described in units of square angstroms (a standard unit of measurement in molecular biology). ASA was first described by Lee & Richards in 1971 and is sometimes called the Lee-Richards molecular surface. ASA is typically calculated using the 'rolling ball' algorithm developed by Shrake & Rupley in 1973. This algorithm uses a sphere (of solvent) of a particular radius to 'probe' the surface of the molecule. Methods of calculating ASA Shrake–Rupley algorithm The Shrake–Rupley algorithm is a numerical method that draws a mesh of points equidistant from each atom of the molecule and uses the number of these points that are solvent accessible to determine the surface area. The points are drawn at a water molecule's estimated radius beyond the van der Waals radius, which is effectively similar to ‘rolli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AMBER
Amber is fossilized tree resin. Examples of it have been appreciated for its color and natural beauty since the Neolithic times, and worked as a gemstone since antiquity."Amber" (2004). In Maxine N. Lurie and Marc Mappen (eds.) ''Encyclopedia of New Jersey'', Rutgers University Press, . Amber is used in jewelry and as a healing agent in Traditional medicine, folk medicine. There are five classes of amber, defined on the basis of their chemical constituents. Because it originates as a soft, sticky tree resin, amber sometimes contains animal and plant material as Inclusion (mineral), inclusions. Amber occurring in coal seams is also called resinite, and the term ''ambrite'' is applied to that found specifically within New Zealand coal seams. Etymology The English word ''amber'' derives from Arabic from Middle Persian 𐭠𐭭𐭡𐭫 (''ʾnbl'' /ambar/, “ambergris”) via Medieval Latin, Middle Latin ''ambar'' and Middle French ''ambre''. The word referred to what is n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Chemistry
Computational chemistry is a branch of chemistry that uses computer simulations to assist in solving chemical problems. It uses methods of theoretical chemistry incorporated into computer programs to calculate the structures and properties of molecules, groups of molecules, and solids. The importance of this subject stems from the fact that, with the exception of some relatively recent findings related to the hydrogen molecular ion (dihydrogen cation), achieving an accurate quantum mechanical depiction of chemical systems analytically, or in a closed form, is not feasible. The complexity inherent in the many-body problem exacerbates the challenge of providing detailed descriptions of quantum mechanical systems. While computational results normally complement information obtained by chemical experiments, it can occasionally predict unobserved chemical phenomena. Overview Computational chemistry differs from theoretical chemistry, which involves a mathematical description of chem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modelling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a molecular mechanics approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a quantum chemistry approach). Molecular mechanics Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical mec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Python (programming Language)
Python is a high-level programming language, high-level, general-purpose programming language. Its design philosophy emphasizes code readability with the use of significant indentation. Python is type system#DYNAMIC, dynamically type-checked and garbage collection (computer science), garbage-collected. It supports multiple programming paradigms, including structured programming, structured (particularly procedural programming, procedural), object-oriented and functional programming. It is often described as a "batteries included" language due to its comprehensive standard library. Guido van Rossum began working on Python in the late 1980s as a successor to the ABC (programming language), ABC programming language, and he first released it in 1991 as Python 0.9.0. Python 2.0 was released in 2000. Python 3.0, released in 2008, was a major revision not completely backward-compatible with earlier versions. Python 2.7.18, released in 2020, was the last release of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
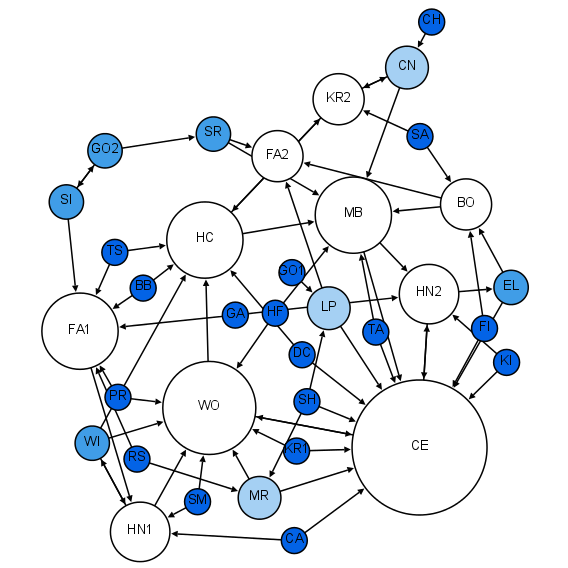
Network Science
Network science is an academic field which studies complex networks such as telecommunication networks, computer networks, biological networks, Cognitive network, cognitive and semantic networks, and social networks, considering distinct elements or actors represented by ''nodes'' (or ''vertices'') and the connections between the elements or actors as ''links'' (or ''edges''). The field draws on theories and methods including graph theory from mathematics, statistical mechanics from physics, data mining and information visualization from computer science, inferential statistics, inferential modeling from statistics, and social structure from sociology. The United States National Research Council defines network science as "the study of network representations of physical, biological, and social phenomena leading to predictive models of these phenomena." Background and history The study of networks has emerged in diverse disciplines as a means of analyzing complex relational ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Journal Of Molecular Graphics
A journal, from the Old French ''journal'' (meaning "daily"), may refer to: *Bullet journal, a method of personal organization *Diary, a record of personal secretive thoughts and as open book to personal therapy or used to feel connected to oneself. A record of what happened over the course of a day or other period *Daybook, also known as a general journal, a daily record of financial transactions *Logbook, a record of events important to the operation of a vehicle, facility, or otherwise *Transaction log, a chronological record of data processing *Travel journal, a record of the traveller's experience during the course of their journey In publishing, ''journal'' can refer to various periodicals or serials: *Academic journal, an academic or scholarly periodical **Scientific journal, an academic journal focusing on science **Medical journal, an academic journal focusing on medicine **Law review, a professional journal focusing on legal interpretation *Magazine, non-academic or scho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Relative Accessible Surface Area
Relative accessible surface area or relative solvent accessibility (RSA) of a protein residue is a measure of residue solvent exposure. It can be calculated by formula: \text = \text / \text where ASA is the solvent accessible surface area and MaxASA is the maximum possible solvent accessible surface area for the residue. Both ASA and MaxASA are commonly measured in ^2 . To measure the relative solvent accessibility of the residue side-chain only, one usually takes MaxASA values that have been obtained from Gly-X-Gly tripeptides, where X is the residue of interest. Several MaxASA scales have been published and are commonly used (see Table). In this table, the more recently published MaxASA values (from Tien et al. 2013) are systematically larger than the older values (from Miller et al. 1987 or Rose et al. 1985). This discrepancy can be traced back to the conformation in which the Gly-X-Gly tripeptides are evaluated to calculate MaxASA. The earlier works used t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
VADAR
Volume, Area, Dihedral Angle Reporter (VADAR) is a freely available protein structure validation web server that was developed as a collaboration between Dr. Brian Sykes and Dr. David Wishart at the University of Alberta. VADAR consists of over 15 different algorithms and programs for assessing and validating peptide and protein structures from their PDB coordinate data. VADAR is capable of determining secondary structure (using three different algorithms), identifying and classifying six different types of beta turns, determining and calculating the strength of C=O -- N-H hydrogen bonds, calculating residue-specific accessible surface areas (ASA), calculating residue volumes, determining backbone and side chain torsion angles (phi, psi, omega and chi angles), assessing local structure quality (through numerous quality indices), evaluating global structure quality, and identifying residue "outliers" (residues with unusual structural features). The results have been validated th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Van Der Waals Surface
The van der Waals surface of a molecule is an abstract representation or model of that molecule, illustrating where, in very rough terms, a surface might reside for the molecule based on the hard cutoffs of van der Waals radii for individual atoms, and it represents a surface through which the molecule might be conceived as interacting with other molecules. Also referred to as a ''van der Waals envelope,'' the van der Waals surface is named for Johannes Diderik van der Waals, a Dutch theoretical physicist and thermodynamicist who developed theory to provide a liquid-gas equation of state that accounted for the non-zero volume of atoms and molecules, and on their exhibiting an attractive force when they interacted (theoretical constructions that also bear his name). van der Waals surfaces are therefore a tool used in the abstract representations of molecules, whether accessed, as they were originally, via hand calculation, or via physical wood/plastic models, or now digitally ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Implicit Solvation
Implicit solvation (sometimes termed continuum solvation) is a method to represent solvent as a continuous medium instead of individual “explicit” solvent molecules, most often used in molecular dynamics simulations and in other applications of molecular mechanics. The method is often applied to estimate free energy of solute-solvent interactions in structural and chemical processes, such as folding or conformational transitions of proteins, DNA, RNA, and polysaccharides, association of biological macromolecules with ligands, or transport of drugs across biological membranes. The implicit solvation model is justified in liquids, where the potential of mean force can be applied to approximate the averaged behavior of many highly dynamic solvent molecules. However, the interfaces and the interiors of biological membranes or proteins can also be considered as media with specific solvation or dielectric properties. These media are not necessarily uniform, since their properties c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frederic Richards
Frederic Middlebrook Richards (August 19, 1925 – January 11, 2009), commonly referred to as Fred Richards, was an American biochemist and biophysics, biophysicist known for solving the pioneering crystallography, crystal structure of the ribonuclease S enzyme in 1967 and for defining the concept of solvent-accessible surface. He contributed many key experimental and theoretical results and developed new methods, garnering over 20,000 journal citations in several quite distinct research areas. In addition to the protein crystallography and biochemistry of ribonuclease S, these included solvent accessibility and internal packing of proteins, the first side-chain rotamer library, high-pressure crystallography, new types of chemical tags such as biotin/avidin, the nuclear magnetic resonance (NMR) chemical shift index, and structural and biophysical characterization of the effects of mutations. Richards spent his entire academic research career at Yale University, where he became S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |