|
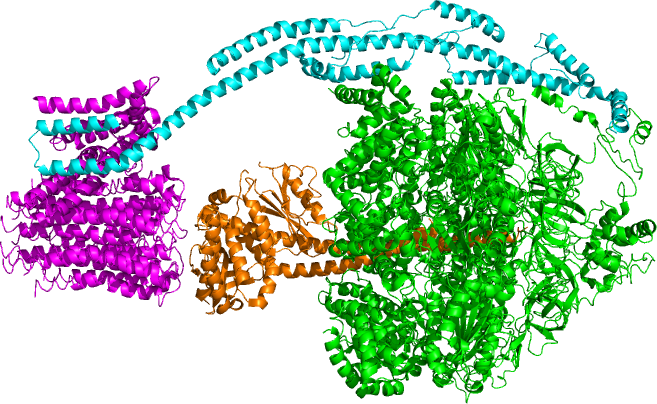
ATP Synthase
ATP synthase is an enzyme that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is: * ADP + Pi + 2H+out ATP + H2O + 2H+in ATP synthase lies across a cellular membrane and forms an aperture that hydron (chemistry), protons can cross from areas of high concentration to areas of low concentration, imparting energy for the synthesis of ATP. This electrochemical gradient is generated by the electron transport chain and allows cells to store energy in ATP for later use. In prokaryote, prokaryotic cells ATP synthase lies across the plasma membrane, while in eukaryote, eukaryotic cells it lies across the inner mitochondrial membrane. Organisms capable of photosynthesis also have ATP synthase across the thylakoid membrane, which in plants is located in the chloroplast and in cyanobacteria is located in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring the angles and intensities of the X-ray diffraction, a crystallography, crystallographer can produce a three-dimensional picture of the density of electrons within the crystal and the positions of the atoms, as well as their chemical bonds, crystallographic disorder, and other information. X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences between various materials, especially minerals and alloys. The method has also revealed the structure and function of many biological molecules, including vitamins, drugs, proteins and nucleic acids such as DNA. X-ray crystall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyanobacteria
Cyanobacteria ( ) are a group of autotrophic gram-negative bacteria that can obtain biological energy via oxygenic photosynthesis. The name "cyanobacteria" () refers to their bluish green (cyan) color, which forms the basis of cyanobacteria's informal common name, blue-green algae. Cyanobacteria are probably the most numerous taxon to have ever existed on Earth and the first organisms known to have produced oxygen, having appeared in the middle Archean eon and apparently originated in a freshwater or terrestrial environment. Their photopigments can absorb the red- and blue-spectrum frequencies of sunlight (thus reflecting a greenish color) to split water molecules into hydrogen ions and oxygen. The hydrogen ions are used to react with carbon dioxide to produce complex organic compounds such as carbohydrates (a process known as carbon fixation), and the oxygen is released as a byproduct. By continuously producing and releasing oxygen over billions of years, cyanobacte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophilic
A hydrophile is a molecule or other molecular entity that is attracted to water molecules and tends to be dissolved by water.Liddell, H.G. & Scott, R. (1940). ''A Greek-English Lexicon'' Oxford: Clarendon Press. In contrast, hydrophobes are not attracted to water and may seem to be repelled by it. Hygroscopics ''are'' attracted to water, but are not dissolved by water. Molecules A hydrophilic molecule or portion of a molecule is one whose interactions with water and other polar substances are more thermodynamically favorable than their interactions with oil or other hydrophobic solvents. They are typically charge-polarized and capable of hydrogen bonding. This makes these molecules soluble not only in water but also in polar solvents. Hydrophilic molecules (and portions of molecules) can be contrasted with hydrophobic molecules (and portions of molecules). In some cases, both hydrophilic and hydrophobic properties occur in a single molecule. An example of these amphiphilic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomycin
Oligomycins are macrolides created by ''Streptomyces'' that are strong antibacterial agents but are often poisonous to other organisms, including humans. Function Oligomycins have use as antibiotics. However, in humans, they have limited or no clinical use due to their toxic effects on mitochondria and ATP synthase. Oligomycin A is an inhibitor of ATP synthase. In oxidative phosphorylation research, it is used to prevent stage 3 (phosphorylating) respiration. Oligomycin A inhibits ATP synthase by blocking its proton channel (FO subunit), which is necessary for oxidative phosphorylation of ADP to ATP (energy production). The inhibition of ATP synthesis by oligomycin A will significantly reduce electron flow through the electron transport chain; however, electron flow is not stopped completely due to a process known as ''proton leak'' or '' mitochondrial uncoupling''. This process is due to facilitated diffusion of protons into the mitochondrial matrix through an uncoupling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphate
Phosphates are the naturally occurring form of the element phosphorus. In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid, phosphoric acid . The phosphate or orthophosphate ion is derived from phosphoric acid by the removal of three protons . Removal of one proton gives the dihydrogen phosphate ion while removal of two protons gives the hydrogen phosphate ion . These names are also used for salts of those anions, such as ammonium dihydrogen phosphate and trisodium phosphate. File:3-phosphoric-acid-3D-balls.png, Phosphoricacid File:2-dihydrogenphosphate-3D-balls.png, Dihydrogenphosphate File:1-hydrogenphosphate-3D-balls.png, Hydrogenphosphate File:0-phosphate-3D-balls.png, Phosphate or orthophosphate In organic chemistry, phosphate or orthophosphate is an organophosphate, an ester of orthophosphoric acid of the form where one ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Diphosphate
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenine attaches to the 1’ carbon. ADP can be interconverted to adenosine triphosphate (ATP) and adenosine monophosphate (AMP). ATP contains one more phosphate group than ADP, while AMP contains one fewer phosphate group. Energy transfer used by all living things is a result of dephosphorylation of ATP by enzymes known as ATPases. The cleavage of a phosphate group from ATP results in the coupling of energy to metabolic reactions and a by-product of ADP. ATP is continually reformed from lower-energy species ADP and AMP. The biosynthesis of ATP is achieved th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known forms of life, it is often referred to as the "molecular unit of currency" for intracellular energy transfer. When consumed in a Metabolism, metabolic process, ATP converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. It is also a Precursor (chemistry), precursor to DNA and RNA, and is used as a coenzyme. An average adult human processes around 50 kilograms (about 100 mole (unit), moles) daily. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of three parts: a sugar, an amine base ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Decomposition
Decomposition is the process by which dead organic substances are broken down into simpler organic or inorganic matter such as carbon dioxide, water, simple sugars and mineral salts. The process is a part of the nutrient cycle and is essential for recycling the finite matter that occupies physical space in the biosphere. Bodies of living organisms begin to decompose shortly after death. Although no two organisms decompose in the same way, they all undergo the same sequential stages of decomposition. Decomposition can be a gradual process for organisms that have extended periods of dormancy. One can differentiate ''abiotic'' decomposition from ''biotic'' decomposition ( biodegradation); the former means "the degradation of a substance by chemical or physical processes", e.g., hydrolysis; the latter means "the metabolic breakdown of materials into simpler components by living organisms", typically by microorganisms. Animals, such as earthworms, also help decompose the organ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. The rate increase occurs because the catalyst allows the reaction to occur by an alternative mechanism which may be much faster than the noncatalyzed mechanism. However the noncatalyzed mechanism does remain possible, so that the total rate (catalyzed plus noncatalyzed) can only increase in the presence of the catalyst and never decrease. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usual ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |