|
ATG8
Autophagy-related protein 8 (Atg8) is a ubiquitin-like protein required for the formation of autophagosomal membranes. The transient conjugation of Atg8 to the autophagosomal membrane through a ubiquitin-like conjugation system is essential for autophagy in eukaryotes. Even though there are homologues in animals (see for example GABARAP, GABARAPL1, GABARAPL2, MAP1LC3A, MAP1LC3B, MAP1LC3B2, and MAP1LC3C), this article mainly focuses on its role in lower eukaryotes such as ''Saccharomyces cerevisiae''. Structure Atg8 is a monomer of 117 aminoacids and a molecular weight of 13,6kDa. It consists of a 5-stranded β-sheet, which is enclosed by two α-helices at one side and one α-helix at the other side and exhibits a conserved GABARAP domain. Even though Atg8 does not show a clear sequence homology to ubiquitin, its crystal structure reveals a conserved ubiquitin-like fold. Function In autophagy Atg8 is one of the key molecular components involved in autophagy, the cellular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-like Protein
Ubiquitin-like proteins (UBLs) are a family of small proteins involved in post-translational modification of other proteins in a cell, usually with a regulatory function. The UBL protein family derives its name from the first member of the class to be discovered, ubiquitin (Ub), best known for its role in regulating protein degradation through covalent modification of other proteins. Following the discovery of ubiquitin, many additional evolutionarily related members of the group were described, involving parallel regulatory processes and similar chemistry. UBLs are involved in a widely varying array of cellular functions including autophagy, protein trafficking, inflammation and immune responses, transcription, DNA repair, RNA splicing, and cellular differentiation. Discovery Ubiquitin itself was first discovered in the 1970s and originally named "ubiquitous immunopoietic polypeptide". Subsequently, other proteins with sequence similarity to ubiquitin were occasionally reported i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAP1LC3B
Microtubule-associated proteins 1A/1B light chain 3B (hereafter referred to as LC3) is a protein that in humans is encoded by the ''MAP1LC3B'' gene. LC3 is a central protein in the autophagy pathway where it functions in autophagy substrate selection and autophagosome biogenesis. LC3 is the most widely used marker of autophagosomes. Discovery LC3 was originally identified as a microtubule associated protein in rat brain. However it was later found that the primary function of LC3 is in autophagy, a process that involves the bulk degradation of cytoplasmic components. The ATG8 protein family MAP1LC3B is a member of the highly conserved ATG8 protein family. ATG8 proteins are present in all known eukaryotic organisms. The animal ATG8 family comprises three subfamilies: (i) microtubule-associated protein 1 light chain 3 (MAP1LC3); (ii) Golgi-associated ATPase enhancer of 16 kDa (GATE-16); and (iii) γ-amino-butyric acid receptor-associate protein (GABARAP). ''MAP1LC3B'' is on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAP1LC3B2
Microtubule-associated proteins 1A/1B light chain 3B (hereafter referred to as LC3) is a protein that in humans is encoded by the ''MAP1LC3B'' gene. LC3 is a central protein in the autophagy pathway where it functions in autophagy substrate selection and autophagosome biogenesis. LC3 is the most widely used marker of autophagosomes. Discovery LC3 was originally identified as a microtubule associated protein in rat brain. However it was later found that the primary function of LC3 is in autophagy, a process that involves the bulk degradation of cytoplasmic components. The ATG8 protein family MAP1LC3B is a member of the highly conserved ATG8 protein family. ATG8 proteins are present in all known eukaryotic organisms. The animal ATG8 family comprises three subfamilies: (i) microtubule-associated protein 1 light chain 3 (MAP1LC3); (ii) Golgi-associated ATPase enhancer of 16 kDa (GATE-16); and (iii) γ-amino-butyric acid receptor-associate protein (GABARAP). ''MAP1LC3B'' is on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macroautophagy
Autophagy (or autophagocytosis; from the Ancient Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components (like mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagy
Autophagy (or autophagocytosis; from the Ancient Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components (like mit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATG12
Autophagy related 12 is a protein that in humans is encoded by the ''ATG12'' gene. Autophagy is a process of bulk protein degradation in which cytoplasmic components, including organelles, are enclosed in double-membrane structures called autophagosomes and delivered to lysosomes or vacuoles for degradation. ATG12 is the human homolog of a yeast protein involved in autophagy (Mizushima et al., 1998). upplied by OMIMref name="entrez"/> Autophagy requires the covalent attachment of the protein Atg12 to ATG5 through a ubiquitin-like conjugation system. The Atg12-Atg5 conjugate then promotes the conjugation of ATG8 to the lipid phosphatidylethanolamine. Atg12 was found to be involved in apoptosis. This protein promotes apoptosis through an interaction with anti-apoptotic members of the Bcl-2 family The Bcl-2 familyTC# 1.A.21 consists of a number of evolutionarily-conserved proteins that share Bcl-2 homology (BH) domains. The Bcl-2 family is most notable for their regulation of apo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAP1LC3A
Microtubule-associated proteins 1A/1B light chain 3A is a protein that in humans is encoded by the ''MAP1LC3A'' gene. Two transcript variants encoding different isoforms have been found for this gene. Function MAP1A and MAP1B are microtubule-associated proteins which mediate the physical interactions between microtubules and components of the cytoskeleton. MAP1A and MAP1B each consist of a heavy chain subunit and multiple light chain subunits. The protein encoded by this gene is one of the light chain subunits and can associate with either MAP1A or MAP1B. MAPLC3A is one of the mammalian homologues of yeast ATG8, an important marker and effector of autophagy. Regulation MAP1LC3A is regulated by several post-translational modifications. These include covalent linkage of the C-terminus to phosphatidylethanolamine in autophagic membranes, and phosphorylation by protein kinase A, which downregulates its autophagy functions. Noncovalent interactions are important for its car ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATG7
Autophagy related 7 is a protein in humans encoded by ''ATG7'' gene. Related to GSA7; APG7L; APG7-LIKE. ATG 7, present in both plant and animal genomes, acts as an essential protein for cell degradation and its recycling. The sequence associates with the ubiquitin- proteasome system, UPS, required for the unique development of an autophagosomal membrane and fusion within cells. ATG7 was identified based on homology to yeast cells ''Pichia pastoris'' GSA7 and ''Saccharomyces cerevisiae'' APG7. The protein appears to be required for fusion of peroxisomal and vacuolar membranes. Autophagy is an important cellular process that helps in maintaining homeostasis. It goes through destroying and recycling the cytoplasmic organelles and macromolecules. During the initiation of autophagy, ATG7 acts like an E-1 enzyme for ubiquitin-like proteins (UBL) such as ATG12 and ATG8. ATG7 helps these UBL proteins in targeting their molecule by binding to them and activating their transfer to an E-2 e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
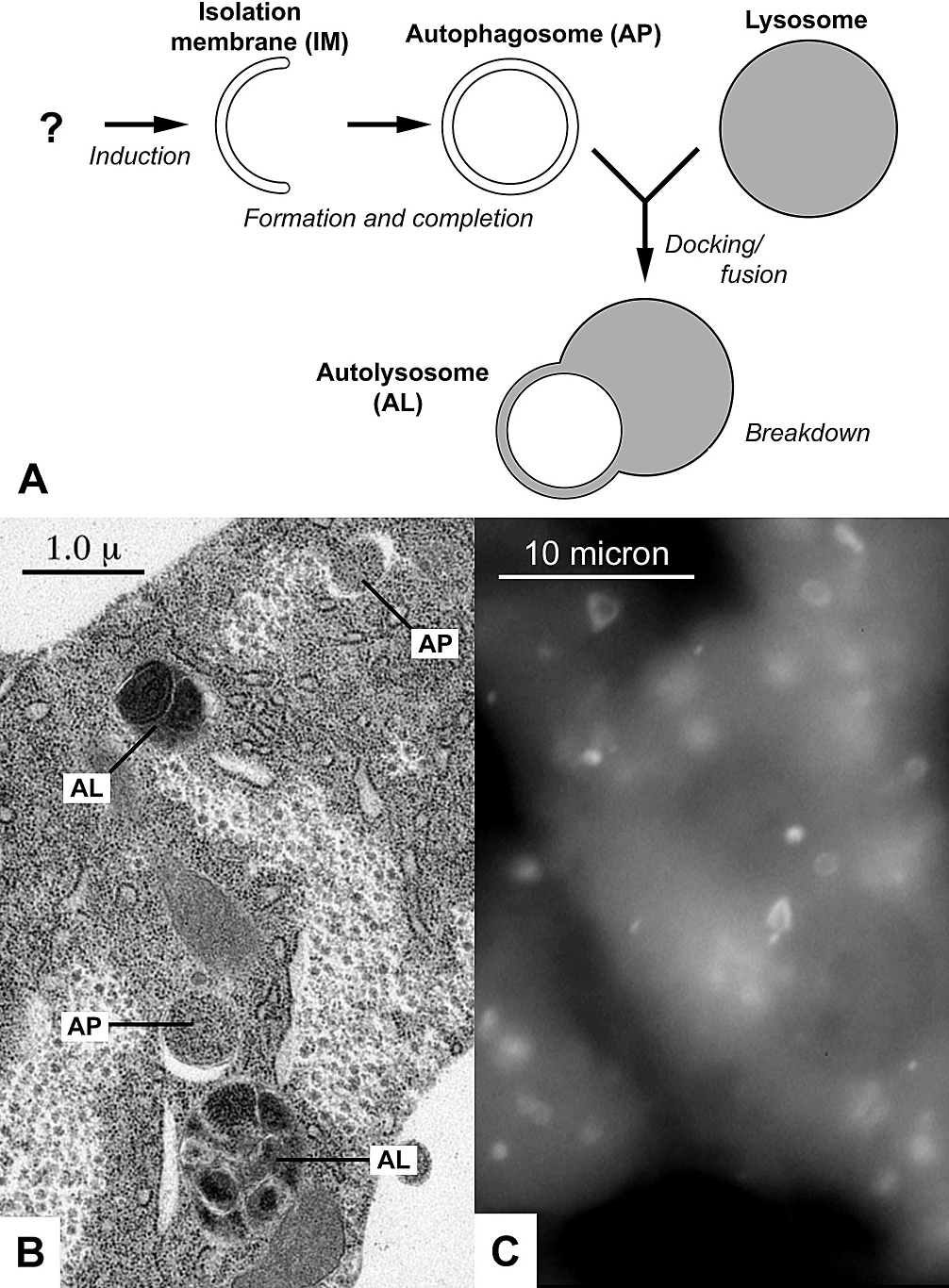
Autolysosome
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents (e.g., abnormal intracellular proteins, excess or damaged organelles, invading microorganisms). After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane. The formation of autophagosomes is regulated by genes that are well-conserved from yeast to higher eukaryotes. The nomenclature of these genes has differed from paper to paper, but it has been simplified in recent years. The gene families formerly known as APG, AUT, CVT, GSA, PAZ, and PDD are now unified as the ATG (AuTophaGy related) family. The size of autophagosomes vary between mammals and yeast. Yeast autophagosomes are about 500-900 nm, while ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E3 Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |