Enhancer-FACS-seq on:
[Wikipedia]
[Google]
[Amazon]
Enhancer-FACS-seq (eFS), developed by the Bulyk lab at Brigham and Women’s Hospital and 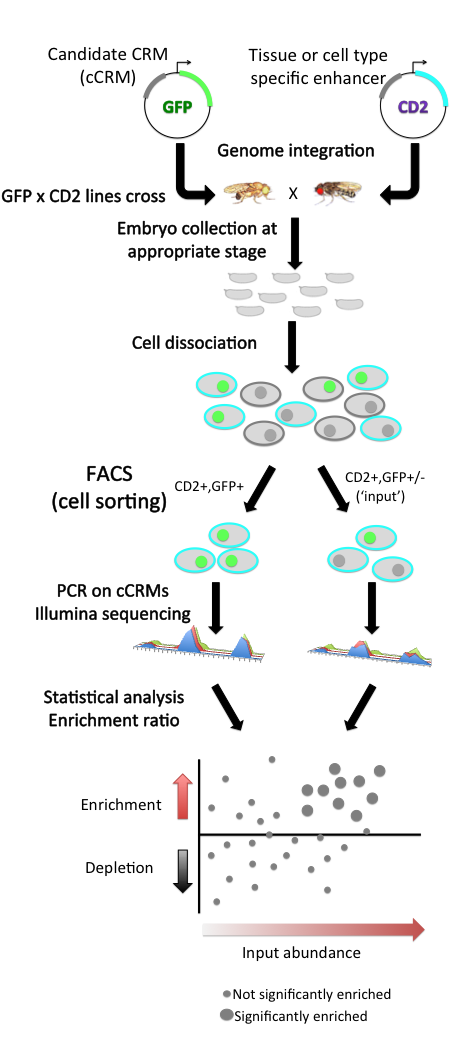
Harvard Medical School
Harvard Medical School (HMS) is the medical school of Harvard University and is located in the Longwood Medical and Academic Area, Longwood Medical Area in Boston, Massachusetts. Founded in 1782, HMS is the third oldest medical school in the Un ...
, is a highly parallel enhancer assay that aims for the identification of active, tissue-specific transcriptional enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, in the context of whole ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly" ...
'' embryos. This technology replaces the use of microscopy to screen for tissue-specific enhancers with fluorescence activated cell sorting ( FACS) of dissociated cells from whole embryos, combined with identification by high-throughput Illumina sequencing.

Introduction
Transcriptional regulation
In metazoans, in order to respond to environmental stress, differentiate properly, and progress normally through the cell cycle, a eukaryotic cell needs a specific and coordinated gene expression program, which involves the highly regulated transcription of thousands of genes. Thisgene regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
is in large part controlled, in a tissue-specific manner, by the binding of transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fun ...
to noncoding genomic regions referred to as cis-regulatory modules (CRMs), activating or repressing gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
by modulating the structure of the chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
and therefore having a positive or negative effect on transcription regulation. CRMs activating gene expression are often referred to as transcriptional enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, whereas those repressing gene expression are referred to as transcriptional silencers.
Enhancer detection in Drosophila melanogaster
Despite being a powerful model organism for biology and the study of transcriptional enhancers, the tissue specific activity of less than 5% of the estimated 50,000 transcriptional enhancers in ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly" ...
'' have been discovered. Over the past decade, the main method for detection of tissue- or cell-type specific activities of enhancers in ''Drosophila melanogaster'' was to test candidate enhancers by traditional reporter assays, which are low-throughput and costly. Over the past few years, even though enhancer discovery has been improved and other parallel reporter assays have been developed, none so far allowed the direct identification of enhancer activity in a genomic
Genomics is an interdisciplinary field of molecular biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, ...
context in cell types of interest in a whole embryo.
Methodology
Each candidate CRM (cCRM) is cloned upstream of a reporter gene. Compared to traditional reporter assays, the main innovation is the use of fluorescence activated cell sorting (FACS) of dissociated cells, instead of microscopy, to screen for tissue-specific enhancers. This approach utilizes a two-marker system: in each embryo, one marker (here, the rat CD2 cell surface protein) is used to label cells of a specific tissue for being sorted by FACS, and the other marker (here, green fluorescent protein GFP) is used as a reporter of CRM activity. Cells are sorted according to their tissue type and then by GFPfluorescence
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with colore ...
, and the cCRMs are recovered by PCR from double-positive sorted cells, and from total input cells. High-throughput sequencing of both populations then allows measuring the relative abundance of each cCRM in input and sorted populations; one can then assess the enrichment or depletion of each cCRM in double-positive cells versus input as a measure of activity in the CD2-positive cell type being tested.
Significant results
In the initial report on this method, a library of ~500 cCRMs was drawn from a variety of genomic data sources (e.g., TF-bound regions, coactivator-bound regions, DNase I hypersensitive sites, and predictions from the Bulyk lab’s PhylCRM algorithm ) by PCR from genomic DNA, and then screened for activity in embryonicmesoderm
The mesoderm is the middle layer of the three germ layers that develops during gastrulation in the very early development of the embryo of most animals. The outer layer is the ectoderm, and the inner layer is the endoderm.Langman's Medical ...
and in specific mesodermal cell types. The results were validated by traditional reporter gene assay in ''Drosophila melanogaster'' embryos for 68 cCRMs tested by eFS. The specificity of eFS was excellent among significantly enriched cCRMs, while sensitivity was good where the majority of the CD2-positive cells express GFP. It was found that the known enhancer-associated chromatin marks H3K27ac, H3K4me1
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono-methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers.
Nomenclature
H3K4me1 i ...
, and Pol II are significantly enriched among the enhancers found to be active in mesoderm
The mesoderm is the middle layer of the three germ layers that develops during gastrulation in the very early development of the embryo of most animals. The outer layer is the ectoderm, and the inner layer is the endoderm.Langman's Medical ...
.
Advantages and future applications
Advantages of eFS
* Highly parallel identification of active, tissue-specific transcriptional enhancers in whole embryos * Candidate enhancers activity assayed in a genomic context * High specificity of detected enhancersFuture applications
The eFS assay could be used to analyze other cell or tissue types. By assessing enrichment in GFP-expressing CD2-negative as well as CD2-positive cells, and by crossing a common pool of reporter transformant male flies to females expressing CD2 in different cell types, it is possible to assay specificity as well as activity. Accelerating the annotation of the regulatory genome in Drosophila should in principle generate the kind of large-scale regulatory interaction data that would allow exploring the network properties of transcriptional regulation.References
{{reflist, 2 Gene expression