DNA-binding domain on:
[Wikipedia]
[Google]
[Amazon]
A DNA-binding domain (DBD) is an independently folded
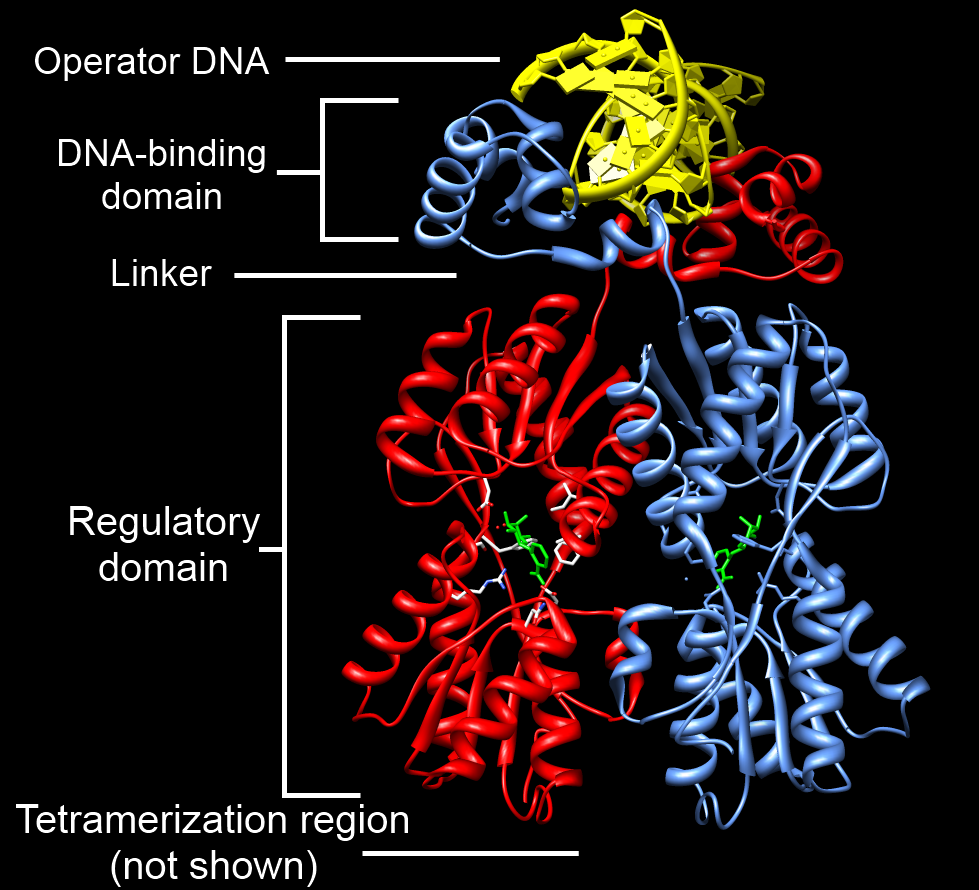 One or more DNA-binding domains are often part of a larger
One or more DNA-binding domains are often part of a larger

.
DBD database of predicted transcription factors
Uses a curated set of DNA-binding domains to predict transcription factors in all completely sequenced genomes
Table of DNA-binding motifs
* * {{MeshName, DNA-Binding+Proteins
DNA-binding domains
in PROSITE Molecular genetics Protein domains it:DNA-binding protein pl:Białko wiążące DNA
protein domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist o ...
that contains at least one structural motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have t ...
that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
Function
 One or more DNA-binding domains are often part of a larger
One or more DNA-binding domains are often part of a larger protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
consisting of further protein domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist o ...
s with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping.
DNA-binding domains with functions involving DNA structure have biological roles in DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inherita ...
, repair, storage
Storage may refer to:
Goods Containers
* Dry cask storage, for storing high-level radioactive waste
* Food storage
* Intermodal container, cargo shipping
* Storage tank
Facilities
* Garage (residential), a storage space normally used to store car ...
, and modification, such as methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
.
Many proteins involved in the regulation of gene expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wid ...
contain DNA-binding domains. For example, proteins that regulate transcription by binding DNA are called transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s. The final output of most cellular signaling cascades is gene regulation.
The DBD interacts with the nucleotides of DNA in a DNA sequence-specific or non-sequence-specific manner, but even non-sequence-specific recognition involves some sort of molecular complementarity between protein and DNA. DNA recognition by the DBD can occur at the major or minor groove of DNA, or at the sugar-phosphate DNA backbone (see the structure of DNA). Each specific type of DNA recognition is tailored to the protein's function. For example, the DNA-cutting enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecule ...
DNAse I
Deoxyribonuclease I (usually called DNase I), is an endonuclease of the DNase family coded by the human gene DNASE1.
DNase I is a nuclease that cleaves DNA preferentially at phosphodiester linkages adjacent to a pyrimidine nucleotide, yielding ...
cuts DNA almost randomly and so must bind to DNA in a non-sequence-specific manner. But, even so, DNAse I recognizes a certain 3-D DNA structure, yielding a somewhat specific DNA cleavage pattern that can be useful for studying DNA recognition by a technique called DNA footprinting.
Many DNA-binding domains must recognize specific DNA sequences, such as DBDs of transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s that activate specific genes, or those of enzymes that modify DNA at specific sites, like restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
s and telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most e ...
. The hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing ...
ing pattern in the DNA major groove is less degenerate than that of the DNA minor groove, providing a more attractive site for sequence
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is called ...
-specific DNA recognition.
The specificity of DNA-binding proteins can be studied using many biochemical and biophysical techniques, such as gel electrophoresis
Gel electrophoresis is a method for separation and analysis of biomacromolecules ( DNA, RNA, proteins, etc.) and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size (IEF ...
, analytical ultracentrifugation, calorimetry, DNA mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
, protein structure
Protein structure is the molecular geometry, three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single ami ...
mutation or modification, nuclear magnetic resonance
Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a strong constant magnetic field are perturbed by a weak oscillating magnetic field (in the near field) and respond by producing an electromagnetic signal with a ...
, x-ray crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angle ...
, surface plasmon resonance, electron paramagnetic resonance, cross-linking and microscale thermophoresis (MST).
DNA-binding protein in genomes
A large fraction of genes in each genome encodes DNA-binding proteins (see Table). However, only a rather small number of protein families are DNA-binding. For instance, more than 2000 of the ~20,000 human proteins are "DNA-binding", including about 750 Zinc-finger proteins.Types
Helix-turn-helix
Originally discovered in bacteria, thehelix-turn-helix
Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove o ...
motif is commonly found in repressor proteins and is about 20 amino acids long. In eukaryotes, the homeodomain comprises 2 helices, one of which recognizes the DNA (aka recognition helix). They are common in proteins that regulate developmental processes ( PROSITEbr>HTH.
Zinc finger
The zinc finger domain is mostly found in eukaryotes, but some examples have been found in bacteria. The zinc finger domain is generally between 23 and 28 amino acids long and is stabilized by coordinating zinc ions with regularly spaced zinc-coordinating residues (either histidines or cysteines). The most common class of zinc finger (Cys2His2) coordinates a single zinc ion and consists of a recognition helix and a 2-strand beta-sheet. In transcription factors these domains are often found in arrays (usually separated by short linker sequences) and adjacent fingers are spaced at 3 basepair intervals when bound to DNA.Leucine zipper
The basic leucine zipper ( bZIP) domain is found mainly in eukaryotes and to a limited extent in bacteria. The bZIP domain contains an alpha helix with a leucine at every 7th amino acid. If two such helices find one another, the leucines can interact as the teeth in a zipper, allowing dimerization of two proteins. When binding to the DNA, basic amino acid residues bind to the sugar-phosphate backbone while the helices sit in the major grooves. It regulates gene expression.Winged helix
Consisting of about 110 amino acids, thewinged helix
Consisting of about 110 amino acids, the domain in winged-helix transcription factors (see Regulation of gene expression) has four helices and a two-strand beta-sheet.
These proteins are classified into 19 families called FoxA-FoxS.
Mutation ...
(WH) domain has four helices and a two-strand beta-sheet.
Winged helix-turn-helix
The wingedhelix-turn-helix
Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove o ...
(wHTH) domain is typically 85-90 amino acids long. It is formed by a 3-helical bundle and a 4-strand beta-sheet (wing).
Helix-loop-helix
Thebasic helix-loop-helix
BASIC (Beginners' All-purpose Symbolic Instruction Code) is a family of general-purpose, high-level programming languages designed for ease of use. The original version was created by John G. Kemeny and Thomas E. Kurtz at Dartmouth College ...
(bHLH) domain is found in some transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s and is characterized by two alpha helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earli ...
(α-helixes) connected by a loop. One helix is typically smaller and due to the flexibility of the loop, allows dimerization by folding and packing against another helix. The larger helix typically contains the DNA-binding regions.
HMG-box
HMG-box domains are found in high mobility group proteins which are involved in a variety of DNA-dependent processes like replication and transcription. They also alter the flexibility of the DNA by inducing bends. The domain consists of three alpha helices separated by loops.Wor3 domain
Wor3 domains, named after the White–Opaque Regulator 3 (Wor3) in ''Candida albicans
''Candida albicans'' is an opportunistic pathogenic yeast that is a common member of the human gut flora. It can also survive outside the human body. It is detected in the gastrointestinal tract and mouth in 40–60% of healthy adults. It is usua ...
'' arose more recently in evolutionary time than most previously described DNA-binding domains and are restricted to a small number of fungi.
OB-fold domain
The OB-fold is a small structural motif originally named for its oligonucleotide/ oligosaccharide binding properties. OB-fold domains range between 70 and 150 amino acids in length. OB-folds bind single-stranded DNA, and hence are single-stranded binding proteins. OB-fold proteins have been identified as critical forDNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inherita ...
, DNA recombination, DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA da ...
, transcription, translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
, cold shock response, and telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature mo ...
maintenance.
Unusual
Immunoglobulin fold
The immunoglobulin domain () consists of a beta-sheet structure with large connecting loops, which serve to recognize either DNA major grooves or antigens. Usually found in immunoglobulin proteins, they are also present in Stat proteins of the cytokine pathway. This is likely because the cytokine pathway evolved relatively recently and has made use of systems that were already functional, rather than creating its own.B3 domain
The B3 DBD (, ) is found exclusively intranscription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s from higher plants and restriction endonucleases EcoRII and BfiI and typically consists of 100-120 residues. It includes seven beta sheets and two alpha helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earli ...
, which form a DNA-binding pseudobarrel protein fold.
TAL effector
TAL effectors are found in bacterial plant pathogens of the genus ''Xanthomonas'' and are involved in regulating the genes of the host plant in order to facilitate bacterial virulence, proliferation, and dissemination. They contain a central region of tandem 33-35 residue repeats and each repeat region encodes a single DNA base in the TALE's binding site. Within the repeat it is residue 13 alone that directly contacts the DNA base, determining sequence specificity, while other positions make contacts with the DNA backbone, stabilising the DNA-binding interaction. Each repeat within the array takes the form of paired alpha-helices, while the whole repeat array forms a right-handed superhelix, wrapping around the DNA-double helix. TAL effector repeat arrays have been shown to contract upon DNA binding and a two-state search mechanism has been proposed whereby the elongated TALE begins to contract around the DNA beginning with a successful Thymine recognition from a unique repeat unit N-terminal of the core TAL-effector repeat array. Related proteins are found in bacterial plant pathogen ''Ralstonia solanacearum'', the fungal endosymbiont ''Burkholderia rhizoxinica'' and two as-yet unidentified marine-microorganisms. The DNA binding code and the structure of the repeat array is conserved between these groups, referred to collectively as the TALE-likes.RNA-guided
The CRISPR/Cas system of ''Streptococcus pyogenes'' can be programmed to direct both activation and repression to natural and artificial eukaryotic promoters through the simple engineering of guide RNAs with base-pairing complementarity to target DNA sites. Cas9 can be used as a customizable RNA-guided DNA-binding platform. Domain Cas9 can be functionalized with regulatory domains of interest (e.g., activation, repression, or epigenetic effector) or with endonuclease domain as a versatile tool for genome engineering biology. and then be targeted to multiple loci using different guide RNAs.See also
* Comparison of nucleic acid simulation softwareReferences
External links
DBD database of predicted transcription factors
Uses a curated set of DNA-binding domains to predict transcription factors in all completely sequenced genomes
Table of DNA-binding motifs
* * {{MeshName, DNA-Binding+Proteins
DNA-binding domains
in PROSITE Molecular genetics Protein domains it:DNA-binding protein pl:Białko wiążące DNA