Comparative genomics on:
[Wikipedia]
[Google]
[Amazon]
 Comparative genomics is a branch of biological research that examines
Comparative genomics is a branch of biological research that examines
 Orthologous sequences are related sequences in different species: a gene exists in the original species, the species divided into two species, so genes in new species are orthologous to the sequence in the original species. Paralogous sequences are separated by gene cloning (gene duplication): if a particular gene in the genome is copied, then the copy of the two sequences is paralogous to the original gene. A pair of orthologous sequences is called orthologous pairs (orthologs), a pair of paralogous sequence is called collateral pairs (paralogs). Orthologous pairs usually have the same or similar function, which is not necessarily the case for collateral pairs. In collateral pairs, the sequences tend to evolve into having different functions.
Comparative genomics exploits both similarities and differences in the
Orthologous sequences are related sequences in different species: a gene exists in the original species, the species divided into two species, so genes in new species are orthologous to the sequence in the original species. Paralogous sequences are separated by gene cloning (gene duplication): if a particular gene in the genome is copied, then the copy of the two sequences is paralogous to the original gene. A pair of orthologous sequences is called orthologous pairs (orthologs), a pair of paralogous sequence is called collateral pairs (paralogs). Orthologous pairs usually have the same or similar function, which is not necessarily the case for collateral pairs. In collateral pairs, the sequences tend to evolve into having different functions.
Comparative genomics exploits both similarities and differences in the
 Comparative genomics starts with basic comparisons of genome size and gene density. For instance, genome size is important for coding capacity and possibly for regulatory reasons. High gene density facilitates genome annotation, analysis of environmental selection. By contrast, low gene density hampers the mapping of genetic disease as in the human genome.
Comparative genomics starts with basic comparisons of genome size and gene density. For instance, genome size is important for coding capacity and possibly for regulatory reasons. High gene density facilitates genome annotation, analysis of environmental selection. By contrast, low gene density hampers the mapping of genetic disease as in the human genome.
heuristic alignment
procedures. Among these, the FASTA and BLAST algorithms are prominent for local pairwise alignment. Recent years have witnessed the development of programs tailored to aligning lengthy sequences, such as MUMmer (1999), BLASTZ (2003), and AVID (2003). While BLASTZ adopts a local approach, MUMmer and AVID are geared towards global alignment. To harness the benefits of both local and global alignment approaches, one effective strategy involves integrating them. Initially, a rapid variant of BLAST known as BLAT is employed to identify homologous "anchor" regions. These anchors are subsequently scrutinized to identify sets exhibiting conserved order and orientation. Such sets of anchors are then subjected to alignment using a global strategy. Additionally, ongoing efforts focus on optimizing existing algorithms to handle the vast amount of genome sequence data by enhancing their speed. Furthermore, MAVID stands out as another noteworthy pairwise alignment program specifically designed for aligning multiple genomes. Pairwise Comparison: The Pairwise comparison of genomic sequence data is widely utilized in comparative gene prediction. Many studies in comparative functional genomics lean on pairwise comparisons, wherein traits of each gene are compared with traits of other genes across species. his method yields many more comparisons than unique observations, making each comparison dependent on others. Multiple comparisons: The comparison of multiple genomes is a natural extension of pairwise inter-specific comparisons. Such comparisons typically aim to identify conserved regions across two phylogenetic scales: 1. Deep comparisons, often referred to as phylogenetic footprinting reveal conservation across higher taxonomic units like vertebrates. 2. Shallow comparisons, recently termed Phylogenetic shadowing, probe conservation across a group of closely related species.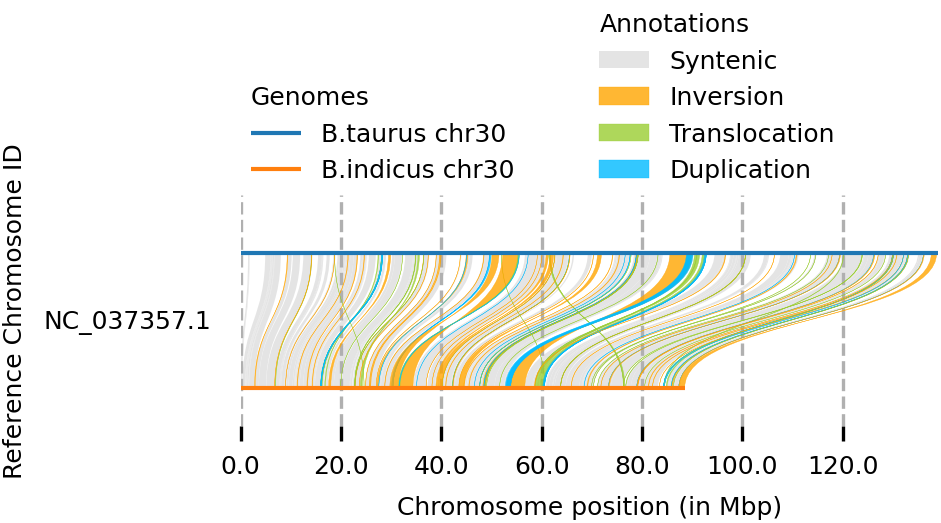



UCSC Browser
'': This site contains the reference sequence and working draft assemblies for a large collection of genomes. *
SyRI
SyRI stands for Synteny and Rearrangement Identifier and is a versatile tool for comparative genomics, offering functionalities for synteny analysis and visualization, aiding in the prediction of genomic differences between related genomes using whole-genome assemblies (WGA). * '
Synmap2
Specifically designed for synteny mapping, Synmap2 efficiently compares genetic maps or assemblies, providing insights into genome evolution and rearrangements among related organisms. * '
GSAlign
GSAlign facilitates accurate alignment of genomic sequences, particularly useful for large-scale comparative genomics studies, enabling researchers to identify similarities and differences across genomes. * '
IGV (Integrative Genomics Viewer)
A widely-used tool for visualizing and analyzing genomic data, IGV supports comparative genomics by enabling users to explore alignments, variants, and annotations across multiple genomes. * '
Manta
Manta is a rapid structural variant caller, crucial for comparative genomics as it detects genomic rearrangements such as insertions, deletions, inversions, and duplications, aiding in understanding genetic variation among populations or species. * '
CNVNatar
CNVNatar specializes in detecting copy number variations (CNVs), which are crucial in understanding genome evolution and population genetics, providing insights into genomic structural changes across different organisms. * '
PIPMaker
PIPMaker facilitates the alignment and comparison of two genomic sequences, enabling the identification of conserved regions, duplications, and evolutionary breakpoints, aiding in comparative genomics analyses. * '
GLASS (Genome-wide Location and Sequence Searcher)
GLASS is a tool for identifying conserved regulatory elements across genomes, crucial for comparative genomics studies focusing on understanding gene regulation and evolution. * PatternHunter: PatternHunter is a versatile tool for sequence analysis, offering functionalities for identifying conserved patterns, motifs, and repeats across genomic sequences, aiding in comparative genomics studies of gene families and regulatory elements. * '
Mummer
Mummer is a suite of tools for whole-genome alignment and comparison, widely used in comparative genomics for identifying similarities, differences, and evolutionary events among genomes at various scales. An advantage of using online tools is that these websites are being developed and updated constantly. There are many new settings and content can be used online to improve efficiency.
Genomes OnLine Database (GOLD)
Genome News Network
JCVI Comprehensive Microbial Resource
Pathema: A Clade Specific Bioinformatics Resource Center
CBS Genome Atlas Database
The UCSC Genome Browser
The U.S. National Human Genome Research Institute
Ensembl
The
Genolevures, comparative genomics of the Hemiascomycetous yeasts
Phylogenetically Inferred Groups (PhIGs)
a recently developed method incorporates phylogenetic signals in building gene clusters for use in comparative genomics.
Metazome
!---->, a resource for the phylogenomic exploration and analysis of Metazoan gene families.
IMG
The Integrated Microbial Genomes system, for comparative genome analysis by the DOE-JGI.
Dcode.org
Dcode.org Comparative Genomics Center.
SUPERFAMILY
Protein annotations for all completely sequenced organisms
Comparative Genomics
Blastology and Open Source: Needs and Deeds
Alignment-free comparative Genomics tool
{{DEFAULTSORT:Comparative Genomics Evolutionary biology Genomics Comparisons
 Comparative genomics is a branch of biological research that examines
Comparative genomics is a branch of biological research that examines genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
sequences across a spectrum of species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
, spanning from humans and mice to a diverse array of organisms from bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
to chimpanzees. This large-scale holistic approach compares two or more genomes to discover the similarities and differences between the genomes and to study the biology
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, History of life, origin, evolution, and ...
of the individual genomes. Comparison of whole genome sequences provides a highly detailed view of how organisms are related to each other at the gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
level. By comparing whole genome sequences, researchers gain insights into genetic relationships between organisms and study evolutionary changes. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
that is evolutionarily conserved between them. Therefore, Comparative genomics provides a powerful tool for studying evolutionary changes among organisms, helping to identify genes that are conserved or common among species, as well as genes that give unique characteristics of each organism. Moreover, these studies can be performed at different levels of the genomes to obtain multiple perspectives about the organisms.
The comparative genomic analysis begins with a simple comparison of the general features of genomes such as genome size, number of genes, and chromosome number. Table 1 presents data on several fully sequenced model organisms, and highlights some striking findings. For instance, while the tiny flowering plant ''Arabidopsis thaliana'' has a smaller genome than that of the fruit fly ''Drosophila melanogaster'' (157 million base pairs v. 165 million base pairs, respectively) it possesses nearly twice as many genes (25,000 v. 13,000). In fact, ''A. thaliana'' has approximately the same number of genes as humans (25,000). Thus, a very early lesson learned in the genomic era is that genome size does not correlate with evolutionary status, nor is the number of genes proportionate to genome size.
In comparative genomics, synteny
In genetics, the term synteny refers to two related concepts:
* In classical genetics, ''synteny'' describes the physical co-localization of genetic loci on the same chromosome within an individual or species.
* In current biology, ''synteny'' m ...
is the preserved order of genes on chromosomes
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most importa ...
of related species indicating their descent from a common ancestor
Common descent is a concept in evolutionary biology applicable when one species is the ancestor of two or more species later in time. According to modern evolutionary biology, all living beings could be descendants of a unique ancestor commonl ...
. Synteny provides a framework in which the conservation of homologous genes and gene order is identified between genomes of different species. Synteny blocks are more formally defined as regions of chromosomes between genomes that share a common order of homologous genes derived from a common ancestor. Alternative names such as conserved synteny or collinearity
In geometry, collinearity of a set of points is the property of their lying on a single line. A set of points with this property is said to be collinear (sometimes spelled as colinear). In greater generality, the term has been used for aligned ...
have been used interchangeably. Comparisons of genome synteny between and within species have provided an opportunity to study evolutionary processes that lead to the diversity of chromosome number and structure in many lineages across the tree of life; early discoveries using such approaches include chromosomal conserved regions in nematodes
The nematodes ( or ; ; ), roundworms or eelworms constitute the phylum Nematoda. Species in the phylum inhabit a broad range of environments. Most species are free-living, feeding on microorganisms, but many are parasitic. Parasitic worms (he ...
and yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
, evolutionary history and phenotypic traits of extremely conserved Hox gene
Hox genes, a subset of homeobox, homeobox genes, are a gene cluster, group of related genes that Evolutionary developmental biology, specify regions of the body plan of an embryo along the craniocaudal axis, head-tail axis of animals. Hox protein ...
clusters across animals and MADS-box
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC /TGG ...
gene family in plants, and karyotype
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by de ...
evolution in mammals and plants.
Furthermore, comparing two genomes not only reveals conserved domains or synteny but also aids in detecting copy number variations
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of G ...
, single nucleotide polymorphisms (SNPs), indels
Indel (insertion-deletion) is a molecular biology term for an insertion or deletion of bases in the genome of an organism. Indels ≥ 50 bases in length are classified as structural variants.
In coding regions of the genome, unless the lengt ...
, and other genomic structural variations.
Virtually started as soon as the whole genomes of two organisms became available (that is, the genomes of the bacteria ''Haemophilus influenzae
''Haemophilus influenzae'' (formerly called Pfeiffer's bacillus or ''Bacillus influenzae'') is a Gram-negative, Motility, non-motile, Coccobacillus, coccobacillary, facultative anaerobic organism, facultatively anaerobic, Capnophile, capnophili ...
'' and ''Mycoplasma genitalium
''Mycoplasma genitalium'' (also known as ''MG','' Mgen, or since 2018, ''Mycoplasmoides genitalium'') is a sexually transmitted, small and pathogenic bacterium that lives on the mucous epithelial cells of the urinary and genital tracts in ...
'') in 1995, comparative genomics is now a standard component of the analysis of every new genome sequence. With the explosion in the number of genome projects
Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism (be it an animal, a plant, a fungus, a bacterium, an archaean, a protist or a virus) and to annotate protein-coding genes and o ...
due to the advancements in DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
technologies, particularly the next-generation sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation ...
methods in late 2000s, this field has become more sophisticated, making it possible to deal with many genomes in a single study. Comparative genomics has revealed high levels of similarity between closely related organisms, such as humans and chimpanzees, and, more surprisingly, similarity between seemingly distantly related organisms, such as humans and the yeast ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
''. It has also showed the extreme diversity of the gene composition in different evolutionary lineages.
History
''See also'': History of genomics Comparative genomics has a root in the comparison ofvirus
A virus is a submicroscopic infectious agent that replicates only inside the living Cell (biology), cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are ...
genomes in the early 1980s. For example, small RNA viruses infecting animals ( picornaviruses) and those infecting plants (cowpea mosaic virus
Cowpea mosaic virus (CPMV) is a non-enveloped plant virus of the comovirus group. Infection of a susceptible cowpea leaf causes a "mosaic" pattern in the leaf, and results in high virus yields (1-2 g/kg). Its genome consists of 2 molecules of ...
) were compared and turned out to share significant sequence similarity and, in part, the order of their genes. In 1986, the first comparative genomic study at a larger scale was published, comparing the genomes of varicella-zoster virus and Epstein-Barr virus that contained more than 100 genes each.
The first complete genome sequence of a cellular organism, that of ''Haemophilus influenzae
''Haemophilus influenzae'' (formerly called Pfeiffer's bacillus or ''Bacillus influenzae'') is a Gram-negative, Motility, non-motile, Coccobacillus, coccobacillary, facultative anaerobic organism, facultatively anaerobic, Capnophile, capnophili ...
'' Rd, was published in 1995. The second genome sequencing paper was of the small parasitic bacterium ''Mycoplasma genitalium'' published in the same year. Starting from this paper, reports on new genomes inevitably became comparative-genomic studies.
''Microbial genomes.'' The first high-resolution whole genome comparison system of microbial genomes of 10-15kbp was developed in 1998 by Art Delcher, Simon Kasif and Steven Salzberg and applied to the comparison of entire highly related microbial organisms with their collaborators at the Institute for Genomic Research (TIGR). The system is called MUMMER
Mummers were bands of men and women from the medieval to early modern era who (during public festivities) dressed in fantastic clothes and costumes and serenaded people outside their houses, or joined the party inside. Costumes were varied and mi ...
and was described in a publication in Nucleic Acids Research in 1999. The system helps researchers to identify large rearrangements, single base mutations, reversals, tandem repeat expansions and other polymorphisms. In bacteria, MUMMER enables the identification of polymorphisms that are responsible for virulence, pathogenicity, and anti-biotic resistance. The system was also applied to the Minimal Organism Project at TIGR and subsequently to many other comparative genomics projects.
''Eukaryote genomes.'' ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
'', the baker's yeast, was the first eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
to have its complete genome sequence published in 1996. After the publication of the roundworm ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''r ...
'' genome in 1998 and together with the fruit fly ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly" ...
'' genome in 2000, Gerald M. Rubin and his team published a paper titled "Comparative Genomics of the Eukaryotes", in which they compared the genomes of the eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
''D. melanogaster'', ''C. elegans'', and ''S. cerevisiae'', as well as the prokaryote
A prokaryote (; less commonly spelled procaryote) is a unicellular organism, single-celled organism whose cell (biology), cell lacks a cell nucleus, nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Gree ...
''H. influenzae''. At the same time, Bonnie Berger, Eric Lander
Eric Steven Lander (born February 3, 1957) is an American mathematician and geneticist who is a professor of biology at the Massachusetts Institute of Technology (MIT), and a professor of systems biology at Harvard Medical School. Eric Lander is ...
, and their team published a paper on whole-genome comparison of human and mouse.
With the publication of the large genomes of vertebrates in the 2000s, including human
Humans (''Homo sapiens'') or modern humans are the most common and widespread species of primate, and the last surviving species of the genus ''Homo''. They are Hominidae, great apes characterized by their Prehistory of nakedness and clothing ...
, the Japanese pufferfish ''Takifugu rubripes
''Takifugu rubripes'', commonly known as the Japanese puffer, Japanese pufferfish, Tiger puffer, or torafugu (), is a pufferfish in the genus '' Takifugu''. It is distinguished by a very small genome that has been fully sequenced because of its ...
'', and mouse
A mouse (: mice) is a small rodent. Characteristically, mice are known to have a pointed snout, small rounded ears, a body-length scaly tail, and a high breeding rate. The best known mouse species is the common house mouse (''Mus musculus'' ...
, precomputed results of large genome comparisons have been released for downloading or for visualization in a genome browser. Instead of undertaking their own analyses, most biologists can access these large cross-species comparisons and avoid the impracticality caused by the size of the genomes.
Next-generation sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation ...
methods, which were first introduced in 2007, have produced an enormous amount of genomic data and have allowed researchers to generate multiple (prokaryotic) draft genome sequences at once. These methods can also quickly uncover single-nucleotide polymorphisms
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in ...
, insertions and deletions by mapping unassembled reads against a well annotated
An annotation is extra information associated with a particular point in a document or other piece of information. It can be a note that includes a comment or explanation. Annotations are sometimes presented in the margin of book pages. For anno ...
reference genome, and thus provide a list of possible gene differences that may be the basis for any functional variation among strains.
Evolutionary principles
One character of biology is evolution,evolutionary theory
Evolution is the change in the heritable characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certai ...
is also the theoretical foundation of comparative genomics, and at the same time the results of comparative genomics unprecedentedly enriched and developed the theory of evolution. When two or more of the genome sequence are compared, one can deduce the evolutionary relationships of the sequences in a phylogenetic tree. Based on a variety of biological genome data and the study of vertical and horizontal evolution processes, one can understand vital parts of the gene structure and its regulatory function.
Similarity of related genomes is the basis of comparative genomics. If two creatures have a recent common ancestor, the differences between the two species genomes are evolved from the ancestors' genome. The closer the relationship between two organisms, the higher the similarities between their genomes. If there is close relationship between them, then their genome will display a linear behaviour (synteny
In genetics, the term synteny refers to two related concepts:
* In classical genetics, ''synteny'' describes the physical co-localization of genetic loci on the same chromosome within an individual or species.
* In current biology, ''synteny'' m ...
), namely some or all of the genetic sequences are conserved. Thus, the genome sequences can be used to identify gene function, by analyzing their homology (sequence similarity) to genes of known function.
 Orthologous sequences are related sequences in different species: a gene exists in the original species, the species divided into two species, so genes in new species are orthologous to the sequence in the original species. Paralogous sequences are separated by gene cloning (gene duplication): if a particular gene in the genome is copied, then the copy of the two sequences is paralogous to the original gene. A pair of orthologous sequences is called orthologous pairs (orthologs), a pair of paralogous sequence is called collateral pairs (paralogs). Orthologous pairs usually have the same or similar function, which is not necessarily the case for collateral pairs. In collateral pairs, the sequences tend to evolve into having different functions.
Comparative genomics exploits both similarities and differences in the
Orthologous sequences are related sequences in different species: a gene exists in the original species, the species divided into two species, so genes in new species are orthologous to the sequence in the original species. Paralogous sequences are separated by gene cloning (gene duplication): if a particular gene in the genome is copied, then the copy of the two sequences is paralogous to the original gene. A pair of orthologous sequences is called orthologous pairs (orthologs), a pair of paralogous sequence is called collateral pairs (paralogs). Orthologous pairs usually have the same or similar function, which is not necessarily the case for collateral pairs. In collateral pairs, the sequences tend to evolve into having different functions.
Comparative genomics exploits both similarities and differences in the proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, re ...
, RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
, and regulatory regions
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the Gene expression, expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living o ...
of different organisms to infer how selection
Selection may refer to:
Science
* Selection (biology), also called natural selection, selection in evolution
** Sex selection, in genetics
** Mate selection, in mating
** Sexual selection in humans, in human sexuality
** Human mating strat ...
has acted upon these elements. Those elements that are responsible for similarities between different species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
should be conserved through time (stabilizing selection
Stabilizing selection (not to be confused with negative or purifying selection) is a type of natural selection in which the population mean stabilizes on a particular non-extreme trait value. This is thought to be the most common mechanism of ...
), while those elements responsible for differences among species should be divergent (positive selection
In population genetics, directional selection is a type of natural selection in which one extreme phenotype is favored over both the other extreme and moderate phenotypes. This genetic selection causes the allele frequency to shift toward the ...
). Finally, those elements that are unimportant to the evolutionary success of the organism will be unconserved (selection is neutral).
One of the important goals of the field is the identification of the mechanisms of eukaryotic genome evolution. It is however often complicated by the multiplicity of events that have taken place throughout the history of individual lineages, leaving only distorted and superimposed traces in the genome of each living organism. For this reason comparative genomics studies of small model organisms
A model organism is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the model organism will provide insight into the workings of other organisms. Mo ...
(for example the model Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''r ...
and closely related Caenorhabditis briggsae) are of great importance to advance our understanding of general mechanisms of evolution.
Role of CNVs in evolution
Comparative genomics plays a crucial role in identifyingcopy number variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of ...
s (CNVs) and understanding their significance in evolution. CNVs, which involve deletions or duplications of large segments of DNA, are recognized as a major source of genetic diversity
Genetic diversity is the total number of genetic characteristics in the genetic makeup of a species. It ranges widely, from the number of species to differences within species, and can be correlated to the span of survival for a species. It is d ...
, influencing gene structure
Gene structure is the organisation of specialised sequence elements within a gene. Genes contain most of the information necessary for living cells to survive and reproduce. In most organisms, genes are made of DNA, where the particular DNA sequen ...
, dosage, and regulation
Regulation is the management of complex systems according to a set of rules and trends. In systems theory, these types of rules exist in various fields of biology and society, but the term has slightly different meanings according to context. Fo ...
. While single nucleotide polymorphisms (SNPs) are more common, CNVs impact larger genomic regions and can have profound effects on phenotype and diversity. Recent studies suggest that CNVs constitute around 4.8–9.5% of the human genome and have a substantial functional and evolutionary impact. In mammals, CNVs contribute significantly to population diversity, influencing gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
and various phenotypic trait
A phenotypic trait, simply trait, or character state is a distinct variant of a phenotypic characteristic of an organism; it may be either inherited or determined environmentally, but typically occurs as a combination of the two.Lawrence, Eleano ...
s. Comparative genomics analyses of human and chimpanzee genomes have revealed that CNVs may play a greater role in evolutionary change compared to single nucleotide changes. Research indicates that CNVs affect more nucleotides than individual base-pair changes, with about 2.7% of the genome affected by CNVs compared to 1.2% by SNPs. Moreover, while many CNVs are shared between humans and chimpanzees, a significant portion is unique to each species. Additionally, CNVs have been associated with genetic diseases
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosome abnormality. Although polygenic disorders are ...
in humans, highlighting their importance in human health. Despite this, many questions about CNVs remain unanswered, including their origin and contributions to evolutionary adaptation and disease. Ongoing research aims to address these questions using techniques like comparative genomic hybridization
Comparative genomic hybridization (CGH) is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The ai ...
, which allows for a detailed examination of CNVs and their significance. When investigators examined the raw sequence data of the human and chimpanzee.
Significance of comparative genomics
Comparative genomics holds profound significance across various fields, including medical research, basic biology, and biodiversity conservation. For instance, in medical research, predicting how genomic variants limited ability to predict which genomic variants lead to changes in organism-level phenotypes, such as increased disease risk in humans, remains challenging due to the immense size of the genome, comprising about three billion nucleotides. To tackle this challenge, comparative genomics offers a solution by pinpointing nucleotide positions that have remained unchanged over millions of years of evolution. These conserved regions indicate potential sites where genetic alterations could have detrimental effects on an organism's fitness, thus guiding the search for disease-causing variants. Moreover, comparative genomics holds promise in unraveling the mechanisms of gene evolution, environmental adaptations, gender-specific differences, and population variations across vertebrate lineages. Furthermore, comparative studies enable the identification of genomic signatures of selection—regions in the genome that have undergone preferential increase and fixation in populations due to their functional significance in specific processes. For instance, in animal genetics, indigenous cattle exhibit superior disease resistance and environmental adaptability but lower productivity compared to exotic breeds. Through comparative genomic analyses, significant genomic signatures responsible for these unique traits can be identified. Using insights from this signature, breeders can make informed decisions to enhance breeding strategies and promote breed development.Methods
Computational approaches are necessary for genome comparisons, given the large amount of data encoded in genomes. Many tools are now publicly available, ranging from whole genome comparisons togene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
analysis. This includes approaches from systems and control, information theory, string analysis and data mining. Computational approaches will remain critical for research and teaching, especially when information science and genome biology is taught in conjunction.
 Comparative genomics starts with basic comparisons of genome size and gene density. For instance, genome size is important for coding capacity and possibly for regulatory reasons. High gene density facilitates genome annotation, analysis of environmental selection. By contrast, low gene density hampers the mapping of genetic disease as in the human genome.
Comparative genomics starts with basic comparisons of genome size and gene density. For instance, genome size is important for coding capacity and possibly for regulatory reasons. High gene density facilitates genome annotation, analysis of environmental selection. By contrast, low gene density hampers the mapping of genetic disease as in the human genome.
Sequence alignment
Alignments are used to capture information about similar sequences such as ancestry, common evolutionary descent, or common structure and function. Alignments can be done for both nucleotide and protein sequences. Alignments consist of local or global pairwise alignments, and multiple sequence alignments. One way to find global alignments is to use a dynamic programming algorithm known as Needleman-Wunsch algorithmheuristic alignment
procedures. Among these, the FASTA and BLAST algorithms are prominent for local pairwise alignment. Recent years have witnessed the development of programs tailored to aligning lengthy sequences, such as MUMmer (1999), BLASTZ (2003), and AVID (2003). While BLASTZ adopts a local approach, MUMmer and AVID are geared towards global alignment. To harness the benefits of both local and global alignment approaches, one effective strategy involves integrating them. Initially, a rapid variant of BLAST known as BLAT is employed to identify homologous "anchor" regions. These anchors are subsequently scrutinized to identify sets exhibiting conserved order and orientation. Such sets of anchors are then subjected to alignment using a global strategy. Additionally, ongoing efforts focus on optimizing existing algorithms to handle the vast amount of genome sequence data by enhancing their speed. Furthermore, MAVID stands out as another noteworthy pairwise alignment program specifically designed for aligning multiple genomes. Pairwise Comparison: The Pairwise comparison of genomic sequence data is widely utilized in comparative gene prediction. Many studies in comparative functional genomics lean on pairwise comparisons, wherein traits of each gene are compared with traits of other genes across species. his method yields many more comparisons than unique observations, making each comparison dependent on others. Multiple comparisons: The comparison of multiple genomes is a natural extension of pairwise inter-specific comparisons. Such comparisons typically aim to identify conserved regions across two phylogenetic scales: 1. Deep comparisons, often referred to as phylogenetic footprinting reveal conservation across higher taxonomic units like vertebrates. 2. Shallow comparisons, recently termed Phylogenetic shadowing, probe conservation across a group of closely related species.

Whole-genome alignment
Whole-genome alignment (WGA) involves predicting evolutionary relationships at the nucleotide level between two or more genomes. It integrates elements of colinear sequence alignment and gene orthology prediction, presenting a greater challenge due to the vast size and intricate nature of whole genomes. Despite its complexity, numerous methods have emerged to tackle this problem because WGAs play a crucial role in various genome-wide analyses, such as phylogenetic inference, genome annotation, and function prediction. Thereby, SyRI (Synteny and Rearrangement Identifier) is one such method that utilizes whole genome alignment and it is designed to identify both structural and sequence differences between two whole-genome assemblies. By taking WGAs as input, SyRI initially scans for disparities in genome structures. Subsequently, it identifies local sequence variations within both rearranged and non-rearranged (syntenic) regions.
Phylogenetic reconstruction
Another computational method for comparative genomics is phylogenetic reconstruction. It is used to describe evolutionary relationships in terms of common ancestors. The relationships are usually represented in a tree called aphylogenetic tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In ...
. Similarly, coalescent theory
Coalescent theory is a Scientific modelling, model of how alleles sampled from a population may have originated from a most recent common ancestor, common ancestor. In the simplest case, coalescent theory assumes no genetic recombination, recombina ...
is a retrospective model to trace alleles of a gene in a population to a single ancestral copy shared by members of the population. This is also known as the most recent common ancestor
A most recent common ancestor (MRCA), also known as a last common ancestor (LCA), is the most recent individual from which all organisms of a set are inferred to have descended. The most recent common ancestor of a higher taxon is generally assu ...
. Analysis based on coalescence theory tries predicting the amount of time between the introduction of a mutation and a particular allele or gene distribution in a population. This time period is equal to how long ago the most recent common ancestor existed. The inheritance relationships are visualized in a form similar to a phylogenetic tree. Coalescence (or the gene genealogy) can be visualized using dendrogram
A dendrogram is a diagram representing a Tree (graph theory), tree graph. This diagrammatic representation is frequently used in different contexts:
* in hierarchical clustering, it illustrates the arrangement of the clusters produced by ...
s.

Genome maps
An additional method in comparative genomics isgenetic mapping
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be sepa ...
. In genetic mapping, visualizing synteny
In genetics, the term synteny refers to two related concepts:
* In classical genetics, ''synteny'' describes the physical co-localization of genetic loci on the same chromosome within an individual or species.
* In current biology, ''synteny'' m ...
is one way to see the preserved order of genes on chromosomes. It is usually used for chromosomes of related species, both of which result from a common ancestor. This and other methods can shed light on evolutionary history. A recent study used comparative genomics to reconstruct 16 ancestral karyotype
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by de ...
s across the mammalian phylogeny. The computational reconstruction showed how chromosomes rearranged themselves during mammal evolution. It gave insight into conservation of select regions often associated with the control of developmental processes. In addition, it helped to provide an understanding of chromosome evolution and genetic diseases
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosome abnormality. Although polygenic disorders are ...
associated with DNA rearrangements.

Tools
Computational tools for analyzing sequences and complete genomes are developing quickly due to the availability of large amount of genomic data. At the same time, comparative analysis tools are progressed and improved. In the challenges about these analyses, it is very important to visualize the comparative results. Visualization of sequence conservation is a tough task of comparative sequence analysis. As we know, it is highly inefficient to examine the alignment of long genomic regions manually. Internet-based genome browsers provide many useful tools for investigating genomic sequences due to integrating all sequence-based biological information on genomic regions. When we extract large amount of relevant biological data, they can be very easy to use and less time-consuming. * 'UCSC Browser
'': This site contains the reference sequence and working draft assemblies for a large collection of genomes. *
Ensembl
Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other v ...
: The Ensembl project produces genome databases for vertebrates and other eukaryotic species, and makes this information freely available online.
* MapView: The Map Viewer provides a wide variety of genome mapping and sequencing data.
* VISTA
Vista may refer to:
Software
*Windows Vista, the line of Microsoft Windows client operating systems released in 2006 and 2007
* VistA, (Veterans Health Information Systems and Technology Architecture) a medical records system of the United States ...
is a comprehensive suite of programs and databases for comparative analysis of genomic sequences. It was built to visualize the results of comparative analysis based on DNA alignments. The presentation of comparative data generated by VISTA can easily suit both small and large scale of data.
* BlueJay Genome Browser: A stand-alone visualization tool for the multi-scale viewing of annotated genomes and other genomic elements.
* 'SyRI
SyRI stands for Synteny and Rearrangement Identifier and is a versatile tool for comparative genomics, offering functionalities for synteny analysis and visualization, aiding in the prediction of genomic differences between related genomes using whole-genome assemblies (WGA). * '
Synmap2
Specifically designed for synteny mapping, Synmap2 efficiently compares genetic maps or assemblies, providing insights into genome evolution and rearrangements among related organisms. * '
GSAlign
GSAlign facilitates accurate alignment of genomic sequences, particularly useful for large-scale comparative genomics studies, enabling researchers to identify similarities and differences across genomes. * '
IGV (Integrative Genomics Viewer)
A widely-used tool for visualizing and analyzing genomic data, IGV supports comparative genomics by enabling users to explore alignments, variants, and annotations across multiple genomes. * '
Manta
Manta is a rapid structural variant caller, crucial for comparative genomics as it detects genomic rearrangements such as insertions, deletions, inversions, and duplications, aiding in understanding genetic variation among populations or species. * '
CNVNatar
CNVNatar specializes in detecting copy number variations (CNVs), which are crucial in understanding genome evolution and population genetics, providing insights into genomic structural changes across different organisms. * '
PIPMaker
PIPMaker facilitates the alignment and comparison of two genomic sequences, enabling the identification of conserved regions, duplications, and evolutionary breakpoints, aiding in comparative genomics analyses. * '
GLASS (Genome-wide Location and Sequence Searcher)
GLASS is a tool for identifying conserved regulatory elements across genomes, crucial for comparative genomics studies focusing on understanding gene regulation and evolution. * PatternHunter: PatternHunter is a versatile tool for sequence analysis, offering functionalities for identifying conserved patterns, motifs, and repeats across genomic sequences, aiding in comparative genomics studies of gene families and regulatory elements. * '
Mummer
Mummer is a suite of tools for whole-genome alignment and comparison, widely used in comparative genomics for identifying similarities, differences, and evolutionary events among genomes at various scales. An advantage of using online tools is that these websites are being developed and updated constantly. There are many new settings and content can be used online to improve efficiency.
Selected applications
Agriculture
Agriculture
Agriculture encompasses crop and livestock production, aquaculture, and forestry for food and non-food products. Agriculture was a key factor in the rise of sedentary human civilization, whereby farming of domesticated species created ...
is a field that reaps the benefits of comparative genomics. Identifying the loci of advantageous genes is a key step in breeding crops that are optimized for greater yield, cost-efficiency, quality, and disease resistance
Disease resistance is the ability to prevent or reduce the presence of diseases in otherwise susceptible hosts. It can arise from genetic or environmental factors, such as incomplete penetrance. Disease tolerance is different as it is the abilit ...
. For example, one genome wide association study conducted on 517 rice landrace
A landrace is a Domestication, domesticated, locally adapted, often traditional variety of a species of animal or plant that has developed over time, through adaptation to its natural and cultural Environment (biophysical), environment of agric ...
s revealed 80 loci associated with several categories of agronomic performance, such as grain weight, amylose
Amylose is a polysaccharide made of α-D-glucose units, bonded to each other through α(1→4) glycosidic bonds. It is one of the two components of starch, making up approximately 20–25% of it. Because of its tightly packed Helix, helical struct ...
content, and drought tolerance
In botany, drought tolerance is the ability by which a plant maintains its biomass production during arid or drought conditions. Some plants are naturally adapted to dry conditions'','' surviving with protection mechanisms such as desiccation tole ...
. Many of the loci were previously uncharacterized. Not only is this methodology powerful, it is also quick. Previous methods of identifying loci associated with agronomic performance required several generations of carefully monitored breeding of parent strains, a time-consuming effort that is unnecessary for comparative genomic studies.
Medicine
Vaccine development
The medical field also benefits from the study of comparative genomics. In an approach known as reverse vaccinology, researchers can discover candidate antigens for vaccine development by analyzing the genome of apathogen
In biology, a pathogen (, "suffering", "passion" and , "producer of"), in the oldest and broadest sense, is any organism or agent that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a Germ theory of d ...
or a family of pathogens. Applying a comparative genomics approach by analyzing the genomes of several related pathogens can lead to the development of vaccines that are multi-protective. A team of researchers employed such an approach to create a universal vaccine for Group B Streptococcus, a group of bacteria responsible for severe neonatal infection. Comparative genomics can also be used to generate specificity for vaccines against pathogens that are closely related to commensal microorganisms. For example, researchers used comparative genomic analysis of commensal
Commensalism is a long-term biological interaction (symbiosis) in which members of one species gain benefits while those of the other species neither benefit nor are harmed. This is in contrast with mutualism, in which both organisms benefit f ...
and pathogenic strains of ''E. coli'' to identify pathogen-specific genes as a basis for finding antigens that result in immune response against pathogenic strains but not commensal ones. In May 2019, using the Global Genome Set, a team in the UK and Australia sequenced thousands of globally-collected isolates of Group A Streptococcus, providing potential targets for developing a vaccine against the pathogen, also known as ''S. pyogenes''.
Personalized Medicine
Personalized Medicine, enabled by Comparative Genomics, represents a revolutionary approach in healthcare, tailoring medical treatment and disease prevention to the individual patient's genetic makeup. By analyzing genetic variation
Genetic variation is the difference in DNA among individuals or the differences between populations among the same species. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources ...
s across populations and comparing them with an individual's genome, clinicians can identify specific genetic marker
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can ...
s associated with disease susceptibility, drug metabolism
Drug metabolism is the metabolic breakdown of drugs by living organisms, usually through specialized enzymatic systems. More generally, xenobiotic metabolism (from the Greek xenos "stranger" and biotic "related to living beings") is the set o ...
, and treatment response. By identifying genetic variants associated with drug metabolism pathways, drug targets, and adverse reaction
An adverse effect is an undesired harmful effect resulting from a medication or other intervention, such as surgery. An adverse effect may be termed a "side effect", when judged to be secondary to a main or therapeutic effect. The term complic ...
s, personalized medicine can optimize medication selection, dosage, and treatment regimens for individual patients. This approach minimizes the risk of adverse drug reactions, enhances treatment efficacy, and improves patient outcomes.
Cancer
Cancer Genomics represents a cutting-edge field within oncology that leverages comparative genomics to revolutionize cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
diagnosis, treatment, and prevention strategies. Comparative genomics plays a crucial role in cancer research by identifying driver mutations, and providing comprehensive analyses of mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s, copy number alterations, structural variants, gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
, and DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
profiles in large-scale studies across different cancer types. By analyzing the genomes of cancer cells and comparing them with healthy cells, researchers can uncover key genetic alterations driving tumorigenesis
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abn ...
, tumor progression, and metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, ...
. This deep understanding of the genomic landscape of cancer has profound implications for precision oncology
Oncology is a branch of medicine that deals with the study, treatment, diagnosis, and prevention of cancer. A medical professional who practices oncology is an ''oncologist''. The name's Etymology, etymological origin is the Greek word ὄγ ...
. Moreover, Comparative Genomics is instrumental in elucidating mechanisms of drug resistance
Drug resistance is the reduction in effectiveness of a medication such as an antimicrobial or an antineoplastic in treating a disease or condition. The term is used in the context of resistance that pathogens or cancers have "acquired", that is ...
—a major challenge in cancer treatment.
Mouse models in immunology
T cells
T cells (also known as T lymphocytes) are an important part of the immune system and play a central role in the adaptive immune response. T cells can be distinguished from other lymphocytes by the presence of a T-cell receptor (TCR) on their ce ...
(also known as a T lymphocytes or a thymocytes) are immune cells
White blood cells (scientific name leukocytes), also called immune cells or immunocytes, are cells of the immune system that are involved in protecting the body against both infectious disease and foreign entities. White blood cells are genera ...
that grow from stem cells in the bone marrow. They assist to defend the body from infection and may aid in the fight against cancer. Because of their morphological, physiological, and genetic resemblance to humans, mice and rats have long been the preferred species for biomedical research animal model
An animal model (short for animal disease model) is a living, non-human, often genetic-engineered animal used during the research and investigation of human disease, for the purpose of better understanding the disease process without the risk of ha ...
s. Comparative Medicine Research is built on the ability to use information from one species to understand the same processes in another. We can get new insights into molecular pathways by comparing human and mouse T cells and their effects on the immune system utilizing comparative genomics. In order to comprehend its TCRs and their genes, Glusman conducted research on the sequencing of the human and mouse T cell receptor loci. TCR genes are well-known and serve as a significant resource for supporting functional genomics and understanding how genes and intergenic regions of the genome contribute to biological processes.
T-cell immune receptors are important in seeing the world of pathogens in the cellular immune system. One of the reasons for sequencing the human and mouse TCR loci was to match the orthologous gene family sequences and discover conserved areas using comparative genomics. These, it was thought, would reflect two sorts of biological information: (1) exons and (2) regulatory sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and vir ...
s. In fact, the majority of V, D, J, and C exons could be identified in this method. The variable regions are encoded by multiple unique DNA elements that are rearranged and connected during T cell (TCR) differentiation: variable (V), diversity (D), and joining (J) elements for the and polypeptides; and V and J elements for the and polypeptides. igure 1However, several short noncoding conserved blocks of the genome had been shown. Both human and mouse motifs are largely clustered in the 200 bp igure 2 the known 3′ enhancers in the TCR/ were identified, and a conserved region of 100 bp in the mouse J intron was subsequently shown to have a regulatory function.
Comparisons of the genomic sequences within each physical site or location of a specific gene on a chromosome (locs) and across species allow for research on other mechanisms and other regulatory signals. Some suggest new hypotheses about the evolution of TCRs, to be tested (and improved) by comparison to the TCR gene complement of other vertebrate species. A comparative genomic investigation of humans and mice will obviously allow for the discovery and annotation of many other genes, as well as identifying in other species for regulatory sequences.
Research
Comparative genomics also opens up new avenues in other areas of research. As DNA sequencing technology has become more accessible, the number of sequenced genomes has grown. With the increasing reservoir of available genomic data, the potency of comparative genomic inference has grown as well. A notable case of this increased potency is found in recentprimate
Primates is an order (biology), order of mammals, which is further divided into the Strepsirrhini, strepsirrhines, which include lemurs, galagos, and Lorisidae, lorisids; and the Haplorhini, haplorhines, which include Tarsiiformes, tarsiers a ...
research. Comparative genomic methods have allowed researchers to gather information about genetic variation
Genetic variation is the difference in DNA among individuals or the differences between populations among the same species. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources ...
, differential gene expression, and evolutionary dynamics in primates that were indiscernible using previous data and methods.
Great Ape Genome Project
The Great Ape Genome Project used comparative genomic methods to investigate genetic variation with reference to the sixgreat ape
The Hominidae (), whose members are known as the great apes or hominids (), are a taxonomic family of primates that includes eight extant species in four genera: '' Pongo'' (the Bornean, Sumatran and Tapanuli orangutan); '' Gorilla'' (the ...
species, finding healthy levels of variation in their gene pool despite shrinking population size. Another study showed that patterns of DNA methylation, which are a known regulation mechanism for gene expression, differ in the prefrontal cortex of humans versus chimps, and implicated this difference in the evolutionary divergence of the two species.
See also
*Data mining
Data mining is the process of extracting and finding patterns in massive data sets involving methods at the intersection of machine learning, statistics, and database systems. Data mining is an interdisciplinary subfield of computer science and ...
* Molecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
* Comparative anatomy
Comparative anatomy is the study of similarities and differences in the anatomy of different species. It is closely related to evolutionary biology and phylogeny (the evolution of species).
The science began in the classical era, continuing in t ...
* Homology
* Sequence mining
Sequential pattern mining is a topic of data mining concerned with finding statistically relevant patterns between data examples where the values are delivered in a sequence. It is usually presumed that the values are discrete, and thus time serie ...
* Alignment-free sequence analysis
References
Further reading
* * * * * * * * * *External links
Genomes OnLine Database (GOLD)
Genome News Network
JCVI Comprehensive Microbial Resource
Pathema: A Clade Specific Bioinformatics Resource Center
CBS Genome Atlas Database
The UCSC Genome Browser
The U.S. National Human Genome Research Institute
Ensembl
The
Ensembl
Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other v ...
Genome Browser
Genolevures, comparative genomics of the Hemiascomycetous yeasts
Phylogenetically Inferred Groups (PhIGs)
a recently developed method incorporates phylogenetic signals in building gene clusters for use in comparative genomics.
Metazome
!---->, a resource for the phylogenomic exploration and analysis of Metazoan gene families.
IMG
The Integrated Microbial Genomes system, for comparative genome analysis by the DOE-JGI.
Dcode.org
Dcode.org Comparative Genomics Center.
SUPERFAMILY
Protein annotations for all completely sequenced organisms
Comparative Genomics
Blastology and Open Source: Needs and Deeds
Alignment-free comparative Genomics tool
{{DEFAULTSORT:Comparative Genomics Evolutionary biology Genomics Comparisons