cDNA library on:
[Wikipedia]
[Google]
[Amazon]
A cDNA library is a combination of cloned cDNA (
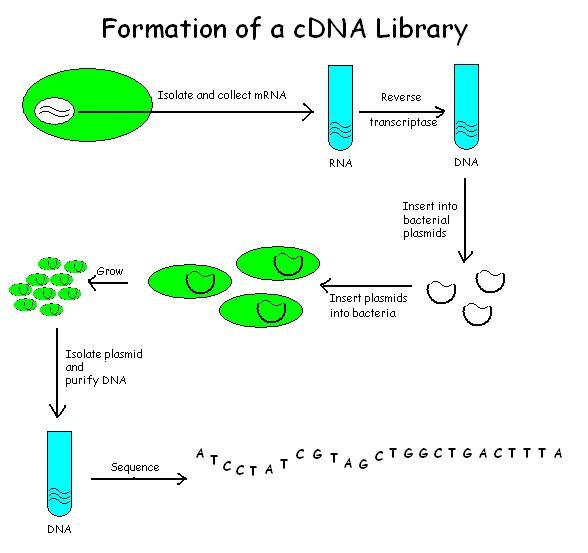 cDNA is created from a mature
cDNA is created from a mature
Functional Annotation of the Mouse database
(
examples of cDNA synthesis and cloning
* Preparation of cDNA libraries for high-throughput RNA sequencing analysis of RNA 5′ ends Molecular biology DNA
complementary DNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
) fragments inserted into a collection of host cells, which constitute some portion of the transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The ...
of the organism and are stored as a "library
A library is a collection of Book, books, and possibly other Document, materials and Media (communication), media, that is accessible for use by its members and members of allied institutions. Libraries provide physical (hard copies) or electron ...
". cDNA is produced from fully transcribed mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue-specific cDNA libraries can be produced. In eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
cells the mature mRNA is already spliced, hence the cDNA produced lacks introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
, and other regulatory elements found in a genomic DNA library.
cDNA Library Construction
 cDNA is created from a mature
cDNA is created from a mature mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
from a eukaryotic cell with the use of reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) distinguishes mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
from tRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
and rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal ...
and can therefore be used as a primer site for reverse transcription. This has the problem that not all transcripts, such as those for the histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
, encode a poly-A tail
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
.
mRNA extraction
Firstly, mRNA template needs to be isolated for the creation of cDNA libraries. Since mRNA only contains exons, the integrity of the isolated mRNA should be considered so that the protein encoded can still be produced. Isolated mRNA should range from 500 bp to 8 kb. Several methods exist for purifying RNA such as trizol extraction and column purification. Column purification can be done using oligomeric dT nucleotide coated resins, and features of mRNA such as having a poly-A tail can be exploited where only mRNA sequences containing said feature will bind. The desired mRNA bound to the column is theneluted
In analytical and organic chemistry, elution is the process of extracting one material from another by washing with a solvent: washing of loaded ion-exchange resins to remove captured ions, or eluting proteins or other biopolymers from an el ...
.
cDNA construction
Once mRNA is purified, an ''oligo-dT'' primer (a short sequence of deoxy-thymidine nucleotides) is bound to the poly-A tail of the RNA. The primer is required to initiate DNA synthesis by the enzymereverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
. This results in the creation of RNA-DNA hybrids where a single strand of complementary DNA is bound to a strand of mRNA. To remove the mRNA, the RNAse H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-nucleotide sequence, sequence-specific endonuclease enzymes that catalysis, catalyze the cleavage of RNA in an RNA/DNA substrate (chemistry), substrate via a hydrolysis, hydrolytic c ...
enzyme is used to cleave the backbone of the mRNA and generate free 3'-OH groups, which is important for the replacement of mRNA with DNA. DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
I is then added, the cleaved RNA acts as a primer the DNA polymerase I can identify and initiate replacement of RNA nucleotides with those of DNA. This is provided by the itself by coiling on itself at the 3' end, generating a '' hairpin loop''. The polymerase extends the 3'-OH end, and later the loop at 3' end is opened by the scissoring action of ''S1 nuclease''. Restriction endonucleases and DNA ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such ...
are then used to clone the sequences into bacterial plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
s.
The cloned bacteria are then selected, commonly through the use of antibiotic selection. Once selected, stocks of the bacteria are created which can later be grown and sequenced to compile the cDNA library.
cDNA Library uses
cDNA libraries are commonly used when reproducing eukaryotic genomes, as the amount of information is reduced to remove the large numbers of non-coding regions from the library. cDNA libraries are used to express eukaryotic genes in prokaryotes. Prokaryotes do not have introns in their DNA and therefore do not possess any enzymes that can cut it out during transcription process. cDNA does not have introns and therefore can be expressed in prokaryotic cells. cDNA libraries are most useful inreverse genetics
Reverse genetics is a method in molecular genetics that is used to help understand the function(s) of a gene by analysing the phenotypic effects caused by genetically engineering specific nucleic acid sequences within the gene. The process proce ...
where the additional genomic information is of less use. Additionally, cDNA libraries are frequently used in functional cloning to identify genes based on the encoded protein's function. When studying eukaryotic DNA, expression libraries are constructed using complementary DNA (cDNA) to help ensure the insert is truly a gene.{{Cite book, title=Biotechnology : applying the genetic revolution, last=P., first=Clark, David, date=2009, publisher=Academic Press/Elsevier, others=Pazdernik, Nanette Jean., isbn=9780121755522, location=Amsterdam, oclc=226038060
cDNA Library vs. Genomic DNA Library
cDNA library lacks the non-coding and regulatory elements found in genomic DNA. Genomic DNA libraries provide more detailed information about the organism, but are more resource-intensive to generate and keep.Cloning of cDNA
cDNA molecules can be cloned by using restriction site linkers. Linkers are short, double stranded pieces of DNA (oligodeoxyribonucleotide) about 8 to 12 nucleotide pairs long that include arestriction endonuclease
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
cleavage site e.g. BamHI. Both the cDNA and the linker have blunt ends which can be ligated together using a high concentration of T4 DNA ligase. Then sticky ends are produced in the cDNA molecule by cleaving the cDNA ends (which now have linkers with an incorporated site) with the appropriate endonuclease. A cloning vector
A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, o ...
(plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
) is then also cleaved with the appropriate endonuclease. Following " sticky end" ligation of the insert into the vector the resulting recombinant DNA molecule is transferred into ''E. coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly foun ...
'' host cell for cloning.
See also
* Functional cloningReferences
External links
Functional Annotation of the Mouse database
(
FANTOM
Fantom is a Swedish velomobile with four wheels, two in the front and two in the rear. It has no front suspension, but has suspension in the rear.
Fantom was never sold as a finished product. Instead it was sold as a set of drawings. The drawin ...
)
examples of cDNA synthesis and cloning
* Preparation of cDNA libraries for high-throughput RNA sequencing analysis of RNA 5′ ends Molecular biology DNA