Bacterial Transcription on:
[Wikipedia]
[Google]
[Amazon]
 Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of
Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of
 The promoter region is a prime regulator of transcription. Promoter regions regulate transcription of all genes within bacteria. As a result of their involvement, the sequence of base pairs within the promoter region is significant; the more similar the promoter region is to the consensus sequence, the tighter RNA polymerase will be able to bind. This binding contributes to the stability of elongation stage of transcription and overall results in more efficient functioning. Additionally, RNA polymerase and σ-factors are in limited supply within any given bacterial cell. Consequently, σ-factor binding to the promoter is affected by these limitations. All promoter regions contain sequences that are considered non-consensus and this helps to distribute σ-factors across the entirety of the genome.
The promoter region is a prime regulator of transcription. Promoter regions regulate transcription of all genes within bacteria. As a result of their involvement, the sequence of base pairs within the promoter region is significant; the more similar the promoter region is to the consensus sequence, the tighter RNA polymerase will be able to bind. This binding contributes to the stability of elongation stage of transcription and overall results in more efficient functioning. Additionally, RNA polymerase and σ-factors are in limited supply within any given bacterial cell. Consequently, σ-factor binding to the promoter is affected by these limitations. All promoter regions contain sequences that are considered non-consensus and this helps to distribute σ-factors across the entirety of the genome.
Bacterial Transcription
– animation
Video animation summarizing the process
{{Transcription Gene expression Bacteria
 Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of
Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
(mRNA) with use of the enzyme RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
.
The process occurs in three main steps: initiation, elongation, and termination; and the end result is a strand of mRNA that is complementary to a single strand of DNA. Generally, the transcribed region accounts for more than one gene. In fact, many prokaryotic genes occur in operons
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splic ...
, which are a series of genes that work together to code for the same protein or gene product and are controlled by a single promoter. Bacterial RNA polymerase is made up of four subunits and when a fifth subunit attaches, called the σ-factor, the polymerase can recognize specific binding sequences in the DNA, called promoters. The binding of the σ-factor to the promoter is the first step in initiation. Once the σ-factor releases from the polymerase, elongation proceeds. The polymerase continues down the double stranded DNA, unwinding it and synthesizing the new mRNA strand until it reaches a termination site. There are two termination mechanisms that are discussed in further detail below. Termination is required at specific sites for proper gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
to occur. Gene expression determines how much gene product, such as protein, is made by the gene. Transcription is carried out by RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
but its specificity is controlled by sequence-specific DNA binding proteins called transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s. Transcription factors work to recognize specific DNA sequences and based on the cells needs, promote or inhibit additional transcription. Similar to other taxa
In biology, a taxon (back-formation from ''taxonomy''; plural taxa) is a group of one or more populations of an organism or organisms seen by taxonomists to form a unit. Although neither is required, a taxon is usually known by a particular nam ...
, bacteria experience bursts of transcription. . NIHMS 816487. EMSID: 85286 The work of the Jones team in Jones et al 2014 explains some of the underlying causes of bursts and other variability, including stability of the resulting mRNA, the strength of promotion encoded in the relevant promoter and the duration of transcription due to strength of the TF binding site. They also found that bacterial TFs linger too briefly for ''TFs'' binding characteristics to explain the sustained transcription of bursts.
Bacterial transcription differs from eukaryotic transcription
Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. ...
in several ways. In bacteria, transcription and translation can occur simultaneously in the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The ...
of the cell, whereas in eukaryotes transcription occurs in the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
* Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
and translation occurs in the cytoplasm. There is only one type of bacterial RNA polymerase whereas eukaryotes have 3 types. Bacteria have a σ-factor that detects and binds to promoter sites but eukaryotes do not need a σ-factor. Instead, eukaryotes have transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The func ...
that allow the recognition and binding of promoter sites.
Overall, transcription within bacteria is a highly regulated process that is controlled by the integration of many signals at a given time. Bacteria heavily rely on transcription and translation to generate proteins that help them respond specifically to their environment.
RNA polymerase
RNA polymerase is composed of a core and a holoenzyme structure. The core enzymes contains the catalytic properties of RNA polymerase and is made up of ββ′α2ω subunits. This sequence is conserved across all bacterial species. The holoenzyme is composed of a specific component known as the sigma factor. The sigma factor functions in aiding in promoter recognition, correct placement of RNA polymerase, and beginning unwinding at the start site. After the sigma factor performs its required function, it dissociates, while the catalytic portion remains on the DNA and continues transcription. Additionally, RNA polymerase contains a core Mg+ ion that assists the enzyme with its catalytic properties. RNA polymerase works by catalyzing the nucleophilic attack of 3’ OH of RNA to the alpha phosphate of a complementary NTP molecule to create a growing strand of RNA from the template strand of DNA. Furthermore, RNA polymerase also displays exonuclease activities, meaning that if improper base pairing is detected, it can cut out the incorrect bases and replace them with the proper, correct one.Initiation
Initiation of transcription requires promoter regions, which are specific nucleotideconsensus sequences
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified r ...
that tell the σ-factor on RNA polymerase where to bind to the DNA. The promoters are usually located 15 to 19 bases apart and are most commonly found upstream of the genes they control. RNA polymerase is made up of 4 subunits, which include two alphas, a beta, and a beta prime (α, α, β, and β'). A fifth subunit, sigma (called the σ-factor), is only present during initiation and detaches prior to elongation. Each subunit plays a role in the initiation of transcription, and the σ-factor ''must'' be present for initiation to occur. When all σ-factor is present, RNA polymerase is in its active form and is referred to as the holoenzyme. When the σ-factor detaches, it is in core polymerase form. The σ-factor recognizes promoter sequences at -35 and -10 regions and transcription begins at the start site (+1). The sequence of the -10 region is TATAAT and the sequence of the -35 region is TTGACA.
* The σ-factor binds to the -35 promoter region. At this point, the holoenzyme is referred to as the ''closed complex'' because the DNA is still double stranded (connected by hydrogen bonds).
*Once the σ-factor binds, the remaining subunits of the polymerase attach to the site. The high concentration of adenine-thymine bonds at the -10 region facilitates the unwinding of the DNA. At this point, the holoenzyme is called the ''open complex''. This open complex is also called the transcription bubble
A transcription bubble is a molecular structure formed during DNA transcription when a limited portion of the DNA double helix is unwound. The size of a transcription bubble ranges from 12-14 base pairs. A transcription bubble is formed when the ...
. Only one strand of DNA, called the template strand (also called the noncoding strand or nonsense/antisense strand), gets transcribed.
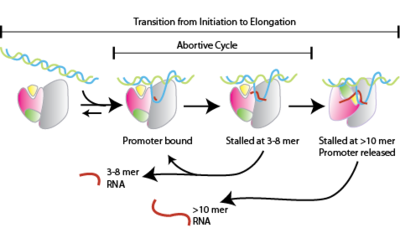
*Transcription begins and short " abortive" nucleotide sequences approximately 10 base pairs long are produced. These short sequences are nonfunctional pieces of RNA that are produced and then released. Generally, this nucleotide sequence consists of about twelve base pairs and aids in contributing to the stability of RNA polymerase so it is able to continue along the strand of DNA.
* The σ-factor is needed to initiate transcription but is not needed to continue transcribing the DNA. The σ-factor dissociates from the core enzyme and elongation proceeds. This signals the end of the initiation phase and the holoenzyme is now in core polymerase form.
 The promoter region is a prime regulator of transcription. Promoter regions regulate transcription of all genes within bacteria. As a result of their involvement, the sequence of base pairs within the promoter region is significant; the more similar the promoter region is to the consensus sequence, the tighter RNA polymerase will be able to bind. This binding contributes to the stability of elongation stage of transcription and overall results in more efficient functioning. Additionally, RNA polymerase and σ-factors are in limited supply within any given bacterial cell. Consequently, σ-factor binding to the promoter is affected by these limitations. All promoter regions contain sequences that are considered non-consensus and this helps to distribute σ-factors across the entirety of the genome.
The promoter region is a prime regulator of transcription. Promoter regions regulate transcription of all genes within bacteria. As a result of their involvement, the sequence of base pairs within the promoter region is significant; the more similar the promoter region is to the consensus sequence, the tighter RNA polymerase will be able to bind. This binding contributes to the stability of elongation stage of transcription and overall results in more efficient functioning. Additionally, RNA polymerase and σ-factors are in limited supply within any given bacterial cell. Consequently, σ-factor binding to the promoter is affected by these limitations. All promoter regions contain sequences that are considered non-consensus and this helps to distribute σ-factors across the entirety of the genome.
Elongation
During elongation, RNA polymerase slides down the double stranded DNA, unwinding it and transcribing (copying) its nucleotide sequence into newly synthesized RNA. The movement of the RNA-DNA complex is essential for thecatalytic
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
mechanism of RNA polymerase. Additionally, RNA polymerase increases the overall stability of this process by acting as a link between the RNA and DNA strands. New nucleotides that are complementary to the DNA template strand are added to the 3' end of the RNA strand. The newly formed RNA strand is ''practically'' identical to the DNA coding strand (sense strand or non-template strand), except it has uracil substituting thymine, and a ribose sugar backbone instead of a deoxyribose sugar backbone. Because nucleoside triphosphates (NTPs) need to attach to the OH- molecule on the 3' end of the RNA, transcription always occurs in the 5' to 3' direction. The four NTPs are adenosine-5'-triphosphate ( ATP), guanoside-5'-triphosphate ( GTP), uridine-5'-triphosphate ( UTP), and cytidine-5'-triphosphate ( CTP). The attachment of NTPs onto the 3' end of the RNA transcript provides the energy required for this synthesis. NTPs are also energy producing molecules that provide the fuel that drives chemical reactions in the cell.
Multiple RNA polymerases can be active at once, meaning many strands of mRNA can be produced very quickly. RNA polymerase moves down the DNA rapidly at approximately 40 bases per second. Due to the quick nature of this process, DNA is continually unwound ahead of RNA polymerase and then rewound once RNA polymerase moves along further. The polymerase has a proofreading mechanism that limits mistakes to about 1 in 10,000 nucleotides transcribed. RNA polymerase has lower fidelity (accuracy) and speed than DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
. DNA polymerase has a very different proofreading mechanism that includes exonuclease activity, which contributes to the higher fidelity. The consequence of an error during RNA synthesis is usually harmless, where as an error in DNA synthesis could be detrimental.
The promoter sequence determines the frequency of transcription of its corresponding gene.
Termination
In order for proper gene expression to occur, transcription must stop at specific sites. Two termination mechanisms are well known: *Intrinsic termination (also called Rho-independent termination): Specific DNA nucleotide sequences signal the RNA polymerase to stop. The sequence is commonly apalindromic sequence
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the comple ...
that causes the strand to loop which stalls the RNA polymerase. Generally, this type of termination follows the same standard procedure. A pause will occur due to a polyuridine sequence that allows the formation of a hairpin loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence wh ...
. This hairpin loop will aid in forming a trapped complex, which will ultimately cause the dissociation of RNA polymerase from the template DNA strand and halt transcription.
*Rho-dependent termination: ρ factor (rho factor) is a terminator protein that attaches to the RNA strand and follows behind the polymerase during elongation. Once the polymerase nears the end of the gene it is transcribing, it encounters a series of G nucleotides which causes it to stall. This stalling allows the rho factor to catch up to the RNA polymerase. The rho protein then pulls the RNA transcript from the DNA template and the newly synthesized mRNA is released, ending transcription. Rho factor is a protein complex that also displays helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separatin ...
activities (is able to unwind the nucleic acid strands). It will bind to the DNA in cytosine rich regions and when RNA polymerase encounters it, a trapped complex will form causing the dissociation of all molecules involved and end transcription.
The termination of DNA transcription in bacteria may be stopped by certain mechanisms wherein the RNA polymerase will ignore the terminator sequence until the next one is reached. This phenomenon is known as antitermination and is utilized by certain bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
s.
References
External links
Bacterial Transcription
– animation
Video animation summarizing the process
{{Transcription Gene expression Bacteria