|
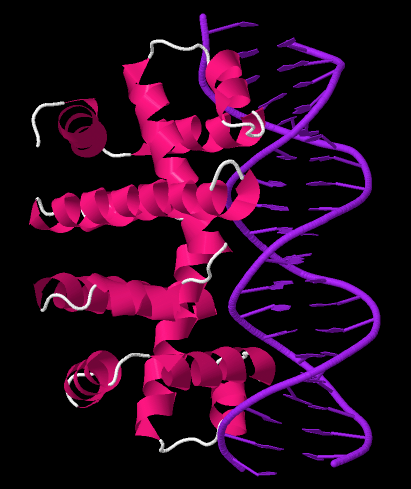
Trp Operon
The ''trp'' operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The ''trp'' operon was first characterized in ''Escherichia coli,'' and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed. The ''trp'' operon contains five structural genes: ''trpE'', ''trpD'', ''trpC'', ''trpB'', and ''trpA'', which encode the enzymes needed to synthesize tryptophan. It also contains a repressive regulator gene called trpR. When tryptophan is present, the trpR protein binds to the operator, blocking transcription of the ''trp'' operon by RNA polymerase. This operon is an example of repressible negative regulation of gene expression. The repressor protein binds to the operator in the presence of tryptophan (repressing transcription) and is released from the operon when tryptophan is absent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, ''Scenedesmus obliquus'', and '' Thra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt wou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypeptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription (biology), transcription of DNA to RNA in the cell's nucleus (cell), nucleus. The entire process is called gene expression. In translation, mRNA, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the Cell (biology), cell. The ribosome facilitates decoding by inducing the binding of Base pair, complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anti-c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expres ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: ''Bacteria'' (formerly Eubacteria) and ''Archaea'' (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes. Besides the absence of a nucleus, prokaryotes also lack mitochondria, or most of the other membrane-bound organelles that characterize the eukaryotic cell. It was once thought that prokaryotic cellular components within the cytop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Attenuator (genetics)
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the ''trp'' operon. Ribosome-mediated attenuation of the ''trp'' operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein. Attenuation is a regulatory feature found throughout Archaea and Bacteria causing premature termination of transcription.Merino E, Yanof ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |