|
Separase
Separase, also known as separin, is a cysteine protease responsible for triggering anaphase by hydrolysing cohesin, which is the protein responsible for binding sister chromatids during the early stage of anaphase. In humans, separin is encoded by the ''ESPL1'' gene. History In ''S. cerevisiae'', separase is encoded by the ''esp1'' gene. Esp1 was discovered by Kim Nasmyth and coworkers in 1998. In 2021, structures of human separase were determined in complex with either securin or CDK1-cyclin B1-CKS1 using cryo-EM by scientists of the University of Geneva. Function Stable cohesion between sister chromatids before anaphase and their timely separation during anaphase are critical for cell division and chromosome inheritance. In vertebrates, sister chromatid cohesion is released in 2 steps via distinct mechanisms. The first step involves phosphorylation of STAG1 or STAG2 in the cohesin complex. The second step involves cleavage of the cohesin subunit SCC1 ( RAD21) by separase, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RAD21
Double-strand-break repair protein rad21 homolog is a protein that in humans is encoded by the ''RAD21'' gene. ''RAD21'' (also known as ''Mcd1'', ''Scc1'', ''KIAA0078'', ''NXP1'', ''HR21''), an essential gene, encodes a DNA double-strand break (DSB) repair protein that is evolutionarily conserved in all eukaryotes from budding yeast to humans. RAD21 protein is a structural component of the highly conserved cohesin complex consisting of RAD21, SMC1A, SMC3, and SCC3 STAG1 (SA1) and STAG2">STAG1.html" ;"title="STAG1">STAG1 (SA1) and STAG2 (SA2) in multicellular organisms] proteins, involved in Establishment of sister chromatid cohesion, sister chromatid cohesion. Discovery ''rad21'' was first cloned by Birkenbihl and Subramani in 1992 by complementing the radiation sensitivity of the ''rad21-45'' mutant fission yeast, ''Schizosaccharomyces pombe'', and the murine and human homologs of ''S. pombe'' rad21 were cloned by McKay, Troelstra, van der Spek, Kanaar, Smit, Hagemeijer, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
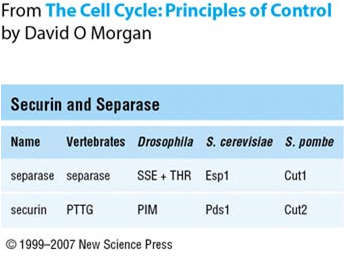
Securin
Securin is a protein involved in control of the metaphase-anaphase transition and anaphase onset. Following bi-orientation of chromosome pairs and inactivation of the spindle checkpoint system, the underlying regulatory system, which includes securin, produces an abrupt stimulus that induces highly synchronous chromosome separation in anaphase. Securin and Separase Securin is initially present in the cytoplasm and binds to separase, a protease that degrades the cohesin rings that link the two sister chromatids. Separase is vital for onset of anaphase. This securin-separase complex is maintained when securin is phosphorylated by Cdk1, inhibiting ubiquitination. When bound to securin, separase is not functional. In addition, both securin and separase are well-conserved proteins (Figure 1). Note that separase cannot function without initially forming the securin-separase complex. This is because securin helps properly fold separase into the functional conformation. However, yeast ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anaphase-promoting Complex
Anaphase-promoting complex (also called the cyclosome or APC/C) is an E3 ubiquitin ligase that marks target cell cycle proteins for degradation by the 26S proteasome. The APC/C is a large complex of 11–13 subunit proteins, including a cullin (Apc2) and RING (Apc11) subunit much like SCF. Other parts of the APC/C have unknown functions but are highly conserved. It was the discovery of the APC/C (and SCF) and their key role in eukaryotic cell-cycle regulation that established the importance of ubiquitin-mediated proteolysis in cell biology. Once perceived as a system exclusively involved in removing damaged protein from the cell, ubiquitination and subsequent protein degradation by the proteasome is now perceived as a universal regulatory mechanism for signal transduction whose importance approaches that of protein phosphorylation. In 2014, the APC/C was mapped in 3D at a resolution of less than a nanometre, which also uncovered its secondary structure. This finding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by S phase of interphase (during which DNA replication occurs) and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other. The process of mitosis is divided into stages corresponding to the completion of one set of activities and the start of the next. These stages are preprophase (specific to plant cells), prophase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complete Network
Network topology is the arrangement of the elements (Data link, links, Node (networking), nodes, etc.) of a communication network. Network topology can be used to define or describe the arrangement of various types of telecommunication networks, including command and control radio networks, industrial Fieldbus, fieldbusses and computer networks. Network topology is the topological structure of a network and may be depicted physically or logically. It is an application of graph theory wherein communicating devices are modeled as nodes and the connections between the devices are modeled as links or lines between the nodes. Physical topology is the placement of the various components of a network (e.g., device location and cable installation), while logical topology illustrates how data flows within a network. Distances between nodes, physical interconnections, Bit rate, transmission rates, or signal types may differ between two different networks, yet their logical topologies may b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cdc14
''Cdc14'' and Cdc14 are a gene and its protein product respectively. Cdc14 is found in most of the eukaryotes. Cdc14 was defined by Hartwell in his famous screen for loci that control the cell cycle of Saccharomyces cerevisiae. Cdc14 was later shown to encode a protein phosphatase. Cdc14 is dual-specificity, which means it has serine/threonine and tyrosine-directed activity. A preference for serines next to proline is reported. Many early studies, especially in the budding yeast Saccharomyces cerevisiae, demonstrated that the protein plays a key role in regulating late mitotic processes. However, more recent work in a range of systems suggests that its cellular function is more complex. Cellular function In Saccharomyces cerevisiae, the species in which Cdc14 activity is best understood and most-studied, the activity of Cdc14 (ScCdc14) leads to mitotic exit by dephosphorylating targets of Cdk1, a well-studied cyclin-dependent protein kinase. Cdc14 antagonizes Cdk1 by stimula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclin-CDK
A cyclin-dependent kinase complex (CDKC, cyclin-CDK) is a protein complex formed by the association of an inactive catalytic subunit of a protein kinase, cyclin-dependent kinase (CDK), with a regulatory subunit, cyclin.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005 Nov;30(11):630-41 Once cyclin-dependent kinases bind to cyclin, the formed complex is in an activated state. Substrate specificity of the activated complex is mainly established by the associated cyclin within the complex. Activity of CDKCs is controlled by phosphorylation of target proteins, as well as binding of inhibitory proteins.Lodish H, Baltimore D, Berk A, Zipursky SL, Matsudaira P, Darnell J. 1995. Molecular Cell Biology. 3rd Ed. New York: Scientific American Books Structure and Regulation The structure of CDKs in complex with a cyclin subunits (CDKC) has long been a goal of structural and cellular biologists starting in the 1990s when the structure of unbou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause disease. Proteolysis can also be used as an analytical tool for studying proteins in the laboratory, and it may also be used in industry, for example in food proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Network Picture
Network, networking and networked may refer to: Science and technology * Network theory, the study of graphs as a representation of relations between discrete objects * Network science, an academic field that studies complex networks Mathematics * Networks, a graph with attributes studied in network theory ** Scale-free network, a network whose degree distribution follows a power law ** Small-world network, a mathematical graph in which most nodes are not neighbors, but have neighbors in common * Flow network, a directed graph where each edge has a capacity and each edge receives a flow Biology * Biological network, any network that applies to biological systems * Ecological network, a representation of interacting species in an ecosystem * Neural network, a network or circuit of neurons Technology and communication * Artificial neural network, a computing system inspired by animal brains * Broadcast network, radio stations, television stations, or other electronic media outlets ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by S phase of interphase (during which DNA replication occurs) and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other. The process of mitosis is divided into stages corresponding to the completion of one set of activities and the start of the next. These stages are preprophase (specific to plant cells), prophase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sister Chromatids
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis ( S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis. Compare sister chromatids to homologous chromosomes, which are the two ''different'' copies of a chromosome that diploid organisms (like humans) inherit, one from each parent. Sister chromatids are by and large identical (since they carry the same alleles, also called variants or versions, of genes) because they derive from one original chromosome. An exception is towards the end of meiosis, after c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |