|
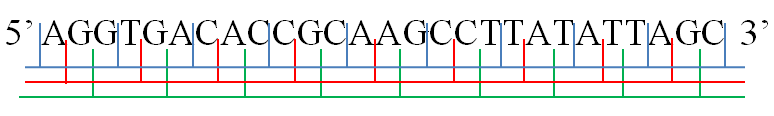
Reading Frame
In molecular biology, a reading frame is a specific choice out of the possible ways to read the nucleic acid sequence, sequence of nucleotides in a nucleic acid (DNA or RNA) molecule as a sequence of triplets. Where these triplets equate to amino acids or stop signals during Translation (biology), translation, they are called codons. A single strand of a nucleic acid molecule has a phosphoryl end, called the 5′-end, and a hydroxyl or 3′-end. These define the Directionality (molecular biology), 5′→3′ direction. There are three reading frames that can be read in this 5′→3′ direction, each beginning from a different nucleotide in a triplet. In a double stranded nucleic acid, an additional three reading frames may be read from the other, Complementarity (molecular biology), complementary strand in the 5′→3′ direction along this strand. As the two strands of a double-stranded nucleic acid molecule are antiparallel, the 5′→3′ direction on the second strand co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the Proteinogenic amino acid, 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups (Alpha and beta carbon, alpha- , beta- , gamma- (γ-) amino acids, etc.); other categories relate to Chemical polarity, polarity, ionization, and side-chain group type (aliphatic, Open-chain compound, acyclic, aromatic, Chemical polarity, polar, etc.). In the form of proteins, amino-acid ''Residue (chemistry)#Biochemistry, residues'' form the second-largest component (water being the largest) of human muscles and other tissue (biology), tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translational Frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses (especially retroviruses), retrotransposons and bacterial insertion elements, and also in some cellular genes. Small molecules, proteins, and nucleic acids have also been found to stimulate levels of frameshifting. In December 2023, it was reported that ''in vitro''-transcribed (IVT) mRNAs in response to BNT162b2 (Pfizer–BioNTech) anti-COVID-19 vaccine caused ribosomal frameshifting. Process overview Proteins are translated by reading tri-nucleotides on the mRNA strand, also known as codons, from one end of the mRNA to the other (from the 5' to the 3' end) starting wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BYDV
Barley yellow dwarf (BYD) is a plant disease caused by the ''barley yellow dwarf virus'' (BYDV), and is the most widely distributed viral disease of cereals. It affects the economically important crop species barley, oats, wheat, maize, triticale and rice. Virus Barley yellow dwarf is caused by barley yellow dwarf viruses. They contain genomes made of ribonucleic acid (RNA). Seven species of barley yellow dwarf virus are recognized, classified as follows: * Kingdom: ''Orthornavirae'' ** Phylum: '' Kitrinoviricota'' *** Class: '' Tolucaviricetes'' **** Order: '' Tolivirales'' ***** Family: ''Tombusviridae'' ****** Genus: '' Luteovirus'' ******* ''Barley yellow dwarf virus kerII'' ******* ''Barley yellow dwarf virus kerIII'' ******* ''Barley yellow dwarf virus MAV'' ******* ''Barley yellow dwarf virus PAS'' ******* ''Barley yellow dwarf virus SGV'' ** Phylum: '' Pisuviricota'' *** Class: '' Pisoniviricetes'' **** Order: '' Sobelivirales'' ***** Family: '' Solemoviridae'' ***** ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hepatitis B Virus
Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus '' Orthohepadnavirus'' and a member of the '' Hepadnaviridae'' family of viruses. This virus causes the disease hepatitis B. Classification Hepatitis B virus is classified in the genus '' Orthohepadnavirus'', which contains 11 other species. The genus is classified as part of the '' Hepadnaviridae'' family, which contains four other genera, '' Avihepadnavirus'', '' Herpetohepadnavirus'', '' Metahepadnavirus'' and '' Parahepadnavirus''. This family of viruses is the only member of the viral order ''Blubervirales''. Viruses similar to hepatitis B have been found in all apes (orangutans, gibbons, bonobos, gorillas and chimpanzees), in Old World monkeys, and in New World woolly monkeys (the woolly monkey hepatitis B virus), suggesting an ancient origin for this virus in primates. The virus is divided into four major serotypes (adr, adw, ayr, ayw) based on antigenic epitopes present on it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Overlapping Gene
An overlapping gene (or OLG) is a gene whose expressible nucleotide sequence partially overlaps with the expressible nucleotide sequence of another gene. In this way, a nucleotide sequence may make a contribution to the function of one or more gene products. Overlapping genes are present in and a fundamental feature of both cellular and viral genomes. The current definition of an overlapping gene varies significantly between eukaryotes, prokaryotes, and viruses. In prokaryotes and viruses overlap must be between coding sequences but not mRNA transcripts, and is defined when these coding sequences share a nucleotide on either the same or opposite strands. In eukaryotes, gene overlap is almost always defined as mRNA transcript overlap. Specifically, a gene overlap in eukaryotes is defined when at least one nucleotide is shared between the boundaries of the primary mRNA transcripts of two or more genes, such that a DNA base mutation at any point of the overlapping region would aff ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homo Sapiens-mtDNA~NC 012920-ATP8+ATP6 Overlap
''Homo'' () is a genus of great ape (family Hominidae) that emerged from the genus ''Australopithecus'' and encompasses only a single extant species, ''Homo sapiens'' (modern humans), along with a number of extinct species (collectively called archaic humans) classified as either ancestral or closely related to modern humans; these include '' Homo erectus'' and '' Homo neanderthalensis''. The oldest member of the genus is '' Homo habilis'', with records of just over 2 million years ago. ''Homo'', together with the genus '' Paranthropus'', is probably most closely related to the species '' Australopithecus africanus'' within ''Australopithecus''.'''' The closest living relatives of ''Homo'' are of the genus '' Pan'' ( chimpanzees and bonobos), with the ancestors of ''Pan'' and ''Homo'' estimated to have diverged around 5.7–11 million years ago during the Late Miocene. ''H. erectus'' appeared about 2 million years ago and spread throughout Africa (debatab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial Genome
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the DNA contained in a eukaryotic cell; most of the DNA is in the cell nucleus, and, in plants and algae, the DNA also is found in plastids, such as chloroplasts. Mitochondrial DNA is responsible for coding of 13 essential subunits of the complex oxidative phosphorylation (OXPHOS) system which has a role in cellular energy conversion. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that human mtDNA has 16,569 base pairs and encodes 13 proteins. As in other vertebrates, the human mitochondrial genetic code differs slightly from nuclear DNA. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It also permits traci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MT-ATP8
''MT-ATP8'' (or ''ATP8'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 8' that encodes a subunit of mitochondrial ATP synthase, ATP synthase Fo subunit 8 (or subunit A6L). This subunit belongs to the Fo complex of the large, transmembrane F-type ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation in the electron transport chain. Specifically, one segment of ATP synthase allows positively charged ions, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Subunit 8 differs in sequence between Metazoa, plants and Fungi. Structure The ATP synthase protein 8 of human and other mammals is encoded in the mitochondrial genome by the ''MT-ATP8'' gene. When the complete human mitochondrial genome was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology, a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, '' Scenedesmus obliquus'', and '' Thraustochytrium''. Reassigned ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Start Codon
The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and archaea and a ''N''-formylmethionine (fMet) in bacteria, mitochondria and plastids. The start codon is often preceded by a 5' untranslated region (5' UTR). In prokaryotes this includes the ribosome binding site. Decoding In all three domains of life, the start codon is decoded by a special "initiation" transfer RNA different from the tRNAs used for elongation. There are important structural differences between an initiating tRNA and an elongating one, with distinguish features serving to satisfy the constraints of the translation system. In bacteria and organelles, an acceptor stem C1:A72 mismatch guide formylation, which directs recruitment by the 30S ribosome into the P site; so-called "3GC" base pairs allow assembly into the 70S ribosome. In eukaryotes and archaea, the T stem prevents the elongation factors from bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |