|
Major Capsid Protein VP1
Major capsid protein VP1 is a viral protein that is the main component of the polyomavirus capsid. VP1 monomers are generally around 350 amino acids long and are capable of self-assembly into an icosahedral structure consisting of 360 VP1 molecules organized into 72 pentamers. VP1 molecules possess a surface binding site that interacts with sialic acids attached to glycans, including some gangliosides, on the surfaces of cells to initiate the process of viral infection. The VP1 protein, along with capsid components VP2 and VP3, is expressed from the "late region" of the circular viral genome. Structure VP1 is the major structural component of the polyomavirus icosahedral capsid, which has T=7 symmetry and a diameter of 40-45 nm. The capsid contains three proteins; VP1 is the primary component and forms a 360-unit outer capsid layer composed of 72 pentamers. The other two components, VP2 and VP3, have high sequence similarity to each other, with VP3 truncated at the N- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Murine Polyomavirus
Murine polyomavirus (also known as mouse polyomavirus, ''Polyomavirus muris'', or ''Mus musculus'' polyomavirus 1, and in older literature as SE polyoma or parotid tumor virus; abbreviated MPyV) is an unenveloped double-stranded DNA virus of the polyomavirus family. The first member of the family discovered, it was originally identified by accident in the 1950s. A component of mouse leukemia extract capable of causing tumors, particularly in the parotid gland, in newborn mice was reported by Ludwik Gross in 1953 and identified as a virus by Sarah Stewart and Bernice Eddy at the National Cancer Institute, after whom it was once called "SE polyoma". Stewart and Eddy would go on to study related polyomaviruses such as SV40 that infect primates, including humans. These discoveries were widely reported at the time and formed the early stages of understanding of oncoviruses. Pathology MPyV is primarily spread among mice via the intranasal route and is shed in urine. Genetic susceptibil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virus-like Particle
Virus-like particles (VLPs) are molecules that closely resemble viruses, but are non-infectious because they contain no viral genetic material. They can be naturally occurring or synthesized through the individual expression of viral structural proteins, which can then self assemble into the virus-like structure. Combinations of structural capsid proteins from different viruses can be used to create recombinant VLPs. Both in-vivo assembly (i.e., assembly inside E. coli bacteria via recombinant co-expression of multiple proteins) and in-vitro assembly (i.e., protein self-assembly in a reaction vessel using stoichiometric quantities of previously purified proteins) have been successfully shown to form virus-like particles. VLPs derived from the Hepatitis B virus (HBV) and composed of the small HBV derived surface antigen (HBsAg) were described in 1968 from patient sera. VLPs have been produced from components of a wide variety of virus families including Parvoviridae (e.g. adeno-associ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
JC Virus
''Human polyomavirus 2'', commonly referred to as the JC virus or John Cunningham virus, is a type of human polyomavirus (formerly known as papovavirus). It was identified by electron microscopy in 1965 by ZuRhein and Chou, and by Silverman and Rubinstein, and later isolated in culture and named using the two initials of a patient, John Cunningham, with progressive multifocal leukoencephalopathy (PML). The virus causes PML and other diseases only in cases of immunodeficiency, as in AIDS or during treatment with immunosuppressive drugs (e.g. in organ transplant patients). Infection and pathogenesis The initial site of infection may be the tonsils, or possibly the gastrointestinal tract. The virus then remains latent in the gastrointestinal tract and can also infect the tubular epithelial cells in the kidneys, where it continues to reproduce, shedding virus particles in the urine. In addition, recent studies suggest that this virus may latently infect the human semen as well as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cluster ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base (chemistry)
In chemistry, there are three definitions in common use of the word base, known as Arrhenius bases, Brønsted bases, and Lewis bases. All definitions agree that bases are substances that react with acids, as originally proposed by G.-F. Rouelle in the mid-18th century. In 1884, Svante Arrhenius proposed that a base is a substance which dissociates in aqueous solution to form Hydroxide ions OH−. These ions can react with hydrogen ions (H+ according to Arrhenius) from the dissociation of acids to form water in an acid–base reaction An acid–base reaction is a chemical reaction that occurs between an acid and a base. It can be used to determine pH via titration. Several theoretical frameworks provide alternative conceptions of the reaction mechanisms and their applica .... A base was therefore a metal hydroxide such as Sodium hydroxide, NaOH or Calcium hydroxide, Ca(OH)2. Such aqueous hydroxide solutions were also described by certain characteristic properties. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disordered Protein
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered protein tertiary structure, three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like Protein aggregation, aggregates, or flexible linkers in large multi-Protein domain, domain proteins. They are sometimes considered as a separate class of proteins along with globular protein, globular, fibrous protein, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent interactions that are highly cooperative and dynamic, lending them importance in Gene expre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
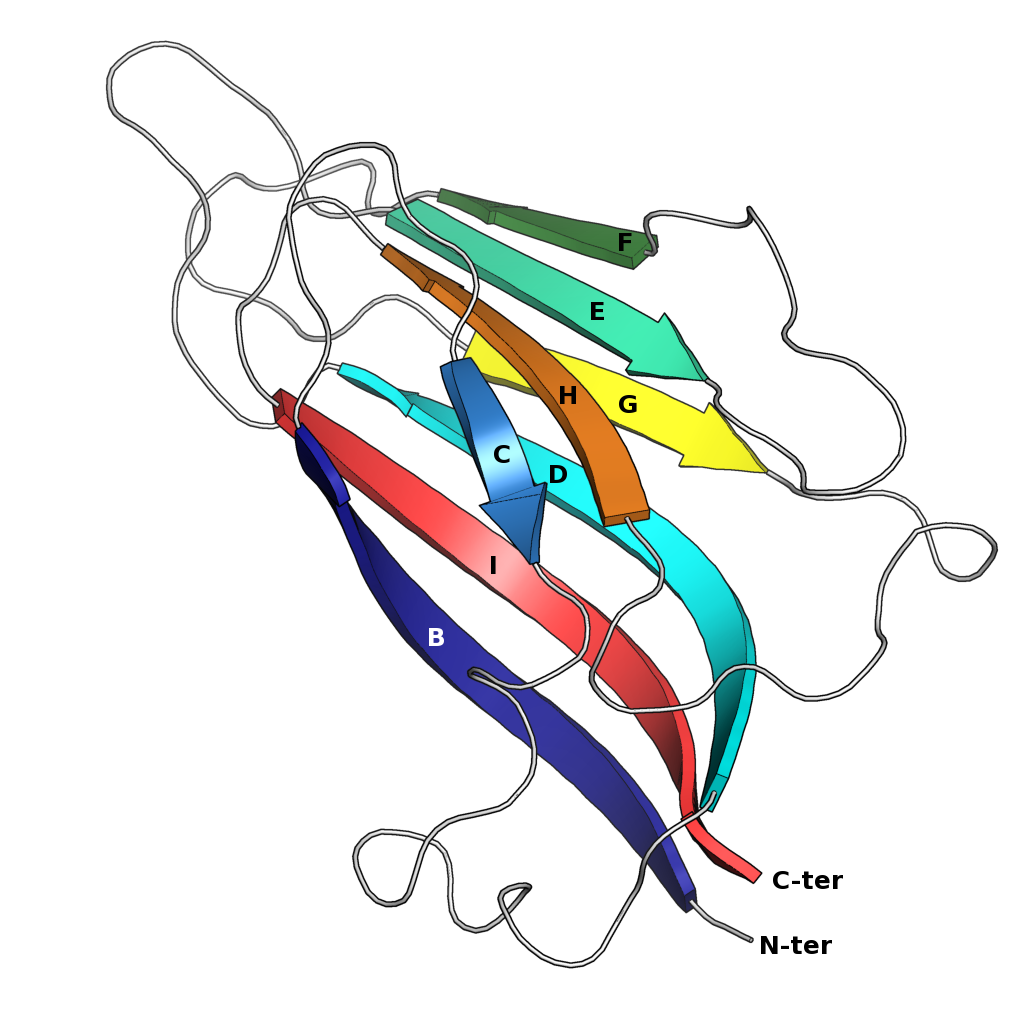
Jelly Roll Fold
The jelly roll or Swiss roll fold is a protein fold or supersecondary structure composed of eight beta strands arranged in two four-stranded sheets. The name of the structure was introduced by Jane S. Richardson in 1981, reflecting its resemblance to the jelly or Swiss roll cake. The fold is an elaboration on the Greek key motif and is sometimes considered a form of beta barrel. It is very common in viral proteins, particularly viral capsid proteins. Taken together, the jelly roll and Greek key structures comprise around 30% of the all-beta proteins annotated in the Structural Classification of Proteins (SCOP) database. Structure The basic jelly roll structure consists of eight beta strands arranged in two four-stranded antiparallel beta sheets which pack together across a hydrophobic interface here citation... uniprot The strands are traditionally labeled B through I for the historical reason that the first solved structure, of a jelly roll capsid protein from the tomato bu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.jpg)