|
Group II Intron
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity (e.g., self- splicing) can occur under high-salt conditions ''in vitro''. However, assistance from proteins is required for ''in vivo'' splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing (see spliceosome) may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or Directed evolution, ''in vitro'' evolved ribozymes are the cleavage (or Ligation (molecular biology), ligation) of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other diazines are pyrazine (nitrogen atoms at the 1 and 4 positions) and pyridazine (nitrogen atoms at the 1 and 2 positions). In nucleic acids, three types of nucleobases are pyrimidine derivatives: cytosine (C), thymine (T), and uracil (U). Occurrence and history The pyrimidine ring system has wide occurrence in nature as substituted and ring fused compounds and derivatives, including the nucleotides cytosine, thymine and uracil, thiamine (vitamin B1) and alloxan. It is also found in many synthetic compounds such as barbiturates and the HIV drug zidovudine. Although pyrimidine derivatives such as alloxan were known in the early 19th century, a laboratory synthesis of a pyrimidine was not carried out until 1879, when Grimaux repor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LtrA
''LtrA'' is an open reading frame found in the '' Lactococcus lactis'' group II introns LtrB. It is an intron-encoded protein, which consists of three subdomains: a reverse-transcriptase/ maturase, DNA endonuclease, and DNA/RNA binding domain In molecular biology, binding domain is a protein domain which binds to a specific atom or molecule, such as calcium or DNA. A protein domain is a part of a protein sequence and a tertiary structure that can change or evolve, function, and live .... LtrA helps to capture and stabilize the catalytically active conformation of the LtrB group II intron RNA. It also functions in group II intron retrohoming. References Prokaryote genes {{molecular-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most eukaryotes and many eukaryotic viruses, and they can be located in both protein-coding genes and genes that function as RNA ( noncoding genes). There are four main types of introns: tRNA introns, group I introns, group I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
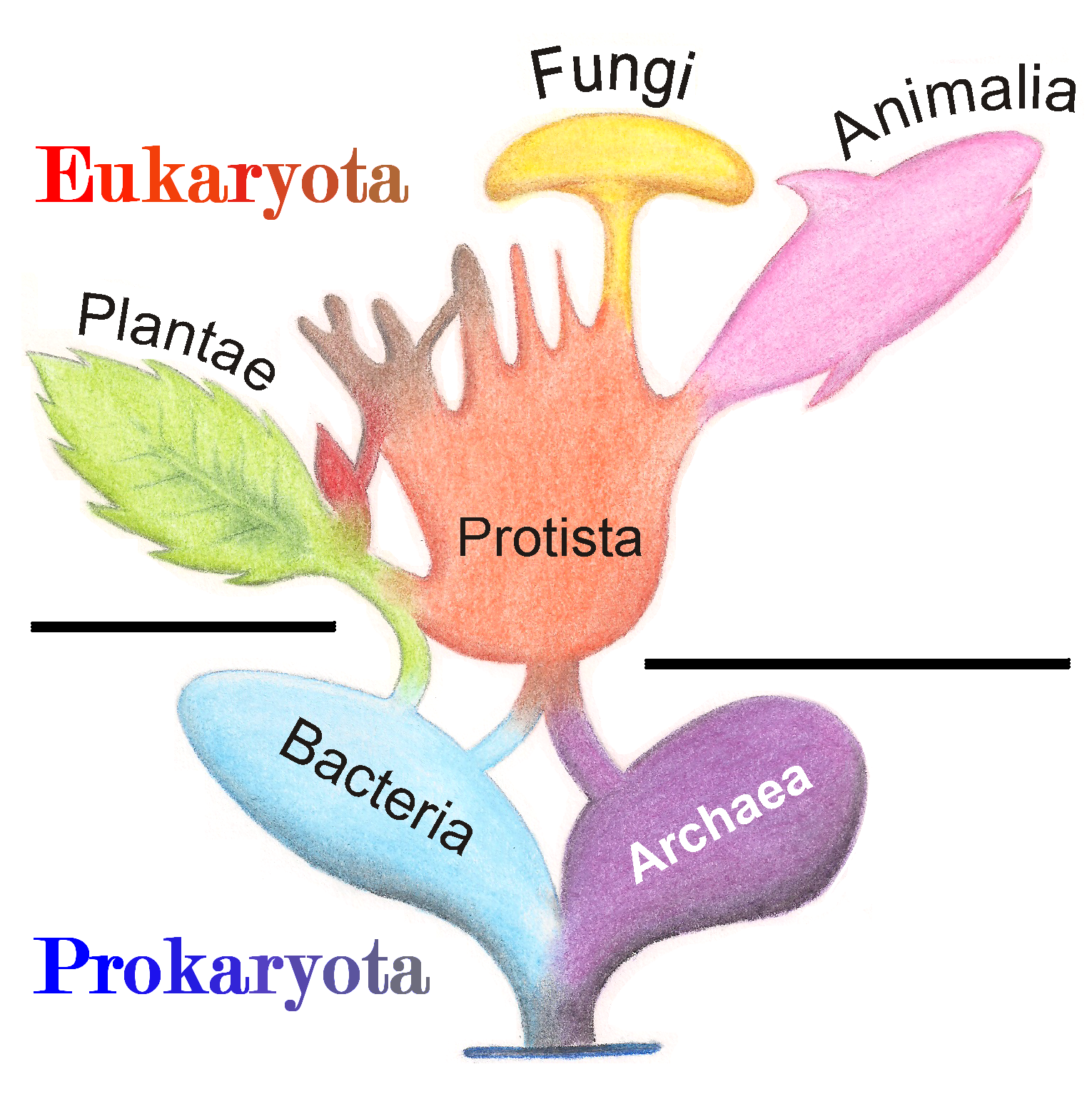
Bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit the air, soil, water, Hot spring, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the nitrogen fixation, fixation of nitrogen from the Earth's atmosphere, atmosphere. The nutrient cycle includes the decomposition of cadaver, dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protists
A protist ( ) or protoctist is any Eukaryote, eukaryotic organism that is not an animal, Embryophyte, land plant, or fungus. Protists do not form a Clade, natural group, or clade, but are a Paraphyly, paraphyletic grouping of all descendants of the last eukaryotic common ancestor excluding land plants, animals, and fungi. Protists were historically regarded as a separate taxonomic rank, taxonomic kingdom (biology), kingdom known as Protista or Protoctista. With the advent of phylogenetic analysis and electron microscopy studies, the use of Protista as a formal taxon was gradually abandoned. In modern classifications, protists are spread across several eukaryotic clades called supergroup (biology), supergroups, such as Archaeplastida (photoautotrophs that includes land plants), SAR supergroup, SAR, Obazoa (which includes fungi and animals), Amoebozoa and "Excavata". Protists represent an extremely large genetic diversity, genetic and ecological diversity in all environments, in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plants
Plants are the eukaryotes that form the kingdom Plantae; they are predominantly photosynthetic. This means that they obtain their energy from sunlight, using chloroplasts derived from endosymbiosis with cyanobacteria to produce sugars from carbon dioxide and water, using the green pigment chlorophyll. Exceptions are parasitic plants that have lost the genes for chlorophyll and photosynthesis, and obtain their energy from other plants or fungi. Most plants are multicellular, except for some green algae. Historically, as in Aristotle's biology, the plant kingdom encompassed all living things that were not animals, and included algae and fungi. Definitions have narrowed since then; current definitions exclude fungi and some of the algae. By the definition used in this article, plants form the clade Viridiplantae (green plants), which consists of the green algae and the embryophytes or land plants ( hornworts, liverworts, mosses, lycophytes, ferns, conifers and other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fungi
A fungus (: fungi , , , or ; or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and mold (fungus), molds, as well as the more familiar mushrooms. These organisms are classified as one of the kingdom (biology)#Six kingdoms (1998), traditional eukaryotic kingdoms, along with Animalia, Plantae, and either Protista or Protozoa and Chromista. A characteristic that places fungi in a different kingdom from plants, bacteria, and some protists is chitin in their cell walls. Fungi, like animals, are heterotrophs; they acquire their food by absorbing dissolved molecules, typically by secreting digestive enzymes into their environment. Fungi do not photosynthesize. Growth is their means of motility, mobility, except for spores (a few of which are flagellated), which may travel through the air or water. Fungi are the principal decomposers in ecological systems. These and other differences place fungi in a single group of related o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Organelle
In cell biology, an organelle is a specialized subunit, usually within a cell (biology), cell, that has a specific function. The name ''organelle'' comes from the idea that these structures are parts of cells, as Organ (anatomy), organs are to the Human body, body, hence ''organelle,'' the suffix ''-elle'' being a diminutive. Organelles are either separately enclosed within their own lipid bilayers (also called membrane-bounded organelles) or are spatially distinct functional units without a surrounding lipid bilayer (non-membrane bounded organelles). Although most organelles are functional units within cells, some functional units that extend outside of cells are often termed organelles, such as cilia, the flagellum and archaellum, and the trichocyst (these could be referred to as membrane bound in the sense that they are attached to (or bound to) the membrane). Organelles are identified by microscopy, and can also be purified by cell fractionation. There are many types of organe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of geneti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code. Overview The process of translation starts with the information stored in the nucleotide sequence of DNA. This is first transformed into mRNA, then tRNA specifies which three-nucleotide codon from the genetic code corresponds to which amino acid. Each mRNA codon is recognized by a particular type of tRNA, which docks to it along ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |