|
Clustal
Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its algorithm are also detailed in their respective categories. Available operating systems listed in the sidebar are a combination of the software availability and may not be supported for every current version of the Clustal tools. Clustal Omega has the widest variety of operating systems out of all the Clustal tools. History There have been many variations of the Clustal software, all of which are listed below: * Clustal: The original software for multiple sequence alignments, created by Des Higgins in 1988, was based on deriving phylogenetic trees from pairwise sequences of amino acids or nucleotides. * ClustalV: The second generation of the Clustal software was released in 1992 and was a rewrite of the original Clustal package. It intr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
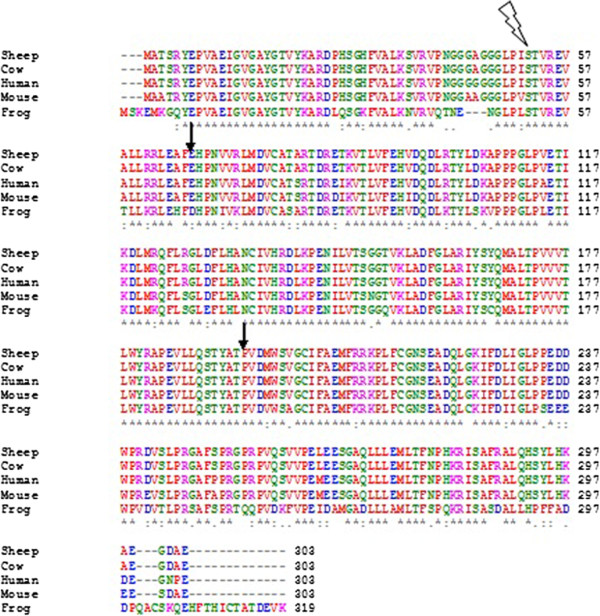
Multiple Sequence Alignment
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Des Higgins
Desmond Gerard Higgins is a Professor of Bioinformatics at University College Dublin, widely known for CLUSTAL,, Broad Institute a series of computer programs for performing multiple sequence alignment. According to ''Nature'', Higgins' papers describing CLUSTAL are among the top ten most highly cited scientific papers of all time. Education Higgins was educated at Trinity College, Dublin where he was awarded a PhD in 1988 for research on numerical taxonomy of Pterygote insects. Research Research in the Higgins laboratory focusses on developing new bioinformatics and statistical tools for evolutionary biology. The CLUSTAL program for multiple sequence alignment has been developed in the Higgins lab and the T-Coffee software was initially developed in the lab with by Cedric Notredame. Multivariate statistics are used to analyse microarray data sets and molecular evolution such as the evolution of promoters, introns and non-coding RNA.Des Higgins Awards and honours Higgin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Toby Gibson
Toby James Gibson is a group leader and biochemist at the European Molecular Biology Laboratory (EMBL) in Heidelberg known for his work on Clustal. According to ''Nature'', Gibson's co-authored papers describing Clustal are among the top ten most highly cited scientific papers of all time. Education Gibson was educated at the University of Edinburgh and went on to his PhD at the University of Cambridge in 1984 on the genome of the Epstein–Barr virus while working in the Medical Research Council (MRC) Laboratory of Molecular Biology (LMB). Career and research Gibson was a postdoctoral research fellow with Sydney Brenner before moving to EMBL in 1986. He was appointed a staff scientist in 1991 and a team leader in 1996 where he has worked since. Gibson’s research interests are in computational biology, bioinformatics, short linear motifs, protein–protein interactions and biological sequence alignment In bioinformatics, a sequence alignment is a way of arranging the seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T-Coffee
T-Coffee (Tree-based Consistency Objective Function for Alignment Evaluation) is a multiple sequence alignment software using a progressive approach. It generates a library of pairwise alignments to guide the multiple sequence alignment. It can also combine multiple sequences alignments obtained previously and in the latest versions can use structural information from PDB files (3D-Coffee). It has advanced features to evaluate the quality of the alignments and some capacity for identifying occurrence of motifs (Mocca). It produces alignment in the aln format (Clustal) by default, but can also produce PIR, MSF, and FASTA format. The most common input formats are supported (FASTA, PIR). Algorithm T-Coffee algorithm consist of two main features, the first by utilizing heterogeneous data sources it is able to provide simple and flexible means of generating multiple alignments. T-coffee can compute multiple alignments using a library that was generated using a mixture of local and glo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
European Bioinformatics Institute
The European Bioinformatics Institute (EMBL-EBI) is an Intergovernmental Organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wellcome Genome Campus in Hinxton near Cambridge, and employs over 600 full-time equivalent (FTE) staff. Institute leaders such as Rolf Apweiler, Alex Bateman, Ewan Birney, and Guy Cochrane, an adviser on the National Genomics Data Center Scientific Advisory Board, serve as part of the international research network of the BIG Data Center at the Beijing Institute of Genomics. Additionally, the EMBL-EBI hosts training programs that teach scientists the fundamentals of the work with biological data and promote the plethora of bioinformatic tools available for their research, both EMBL-EBI and non-EMBL-EBI-based. Bioinformatic services One of the roles of the EMBL-EBI is to index and maintain biological data in a set of databases, including E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Desmond G
Desmond or Desmond's may refer to: Arts and entertainment * ''Desmond'' (novel), 1792 novel by Charlotte Turner Smith * ''Desmond's'', 1990s British television sitcom Ireland * Kingdom of Desmond, medieval Irish kingdom * Earl of Desmond, Irish aristocratic title * Desmond Rebellions, Irish rebellions during the 16th century led by the Earl of Desmond Science and technology * DESMOND (diabetes) (Diabetes Education and Self Management for Ongoing and Newly Diagnosed), a UK NHS diabetes education programme * Desmond (software), molecular dynamics simulation software * Storm Desmond, a windstorm in Britain and Ireland in 2015 Other uses * Desmond (name), a common given name and surname * Desmond (horse) (1896-1913), Thoroughbred racehorse * Desmond's (department store), a former US store * Desmond, slang term for the British 2:2 degree classification See also * Desman, a tribe of aquatic mammals * Clíodhna, principal goddess of Desmond, or South Munster * Limerick Desmond League ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Phylogenetics
Molecular phylogenetics () is the branch of phylogeny that analyzes genetic, hereditary molecular differences, predominantly in DNA sequences, to gain information on an organism's evolutionary relationships. From these analyses, it is possible to determine the processes by which diversity among species has been achieved. The result of a molecular phylogenetic analysis is expressed in a phylogenetic tree. Molecular phylogenetics is one aspect of molecular systematics, a broader term that also includes the use of molecular data in taxonomy and biogeography. Molecular phylogenetics and molecular evolution correlate. Molecular evolution is the process of selective changes (mutations) at a molecular level (genes, proteins, etc.) throughout various branches in the tree of life (evolution). Molecular phylogenetics makes inferences of the evolutionary relationships that arise due to molecular evolution and results in the construction of a phylogenetic tree. History The theoretical frame ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X Window System
The X Window System (X11, or simply X) is a windowing system for bitmap displays, common on Unix-like operating systems. X provides the basic framework for a GUI environment: drawing and moving windows on the display device and interacting with a mouse and keyboard. X does not mandate the user interfacethis is handled by individual programs. As such, the visual styling of X-based environments varies greatly; different programs may present radically different interfaces. X originated as part of Project Athena at Massachusetts Institute of Technology (MIT) in 1984. The X protocol has been at version 11 (hence "X11") since September 1987. The X.Org Foundation leads the X project, with the current reference implementation, X.Org Server, available as free and open-source software under the MIT License and similar permissive licenses. Purpose and abilities X is an architecture-independent system for remote graphical user interfaces and input device capabilities. Each person using a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA Format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences. The format originates from the FASTA software package, but has now become a near universal standard in the field of bioinformatics. The simplicity of FASTA format makes it easy to manipulate and parse sequences using text-processing tools and scripting languages like the R programming language, Python, Ruby, Haskell, and Perl. Original format & overview The original FASTA/Pearson format is described in the documentation for the FASTA suite of programs. It can be downloaded with any free distribution of FASTA (see fasta20.doc, fastaVN.doc or fastaVN.me—where VN is the Version Number). In the original format, a sequence was represented as a series of lines, each of whic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Information Resource
The Protein Information Resource (PIR), located at Georgetown University Medical Center, is an integrated public bioinformatics resource to support genomic and proteomic research, and scientific studies. It contains protein sequences databases History PIR was established in 1984 by the National Biomedical Research Foundation as a resource to assist researchers and customers in the identification and interpretation of protein sequence information. Prior to that, the foundation compiled the first comprehensive collection of macromolecular sequences in the Atlas of Protein Sequence and Structure, published from 1964 to 1974 under the editorship of Margaret Dayhoff. Dayhoff and her research group pioneered in the development of computer methods for the comparison of protein sequences, for the detection of distantly related sequences and duplications within sequences, and for the inference of evolutionary histories from alignments of protein sequences. Winona Barker and Robert Ledley a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |