|
Autophosphorylation
Autophosphorylation is a type of post-translational modification of proteins. It is generally defined as the phosphorylation of the kinase by itself. In eukaryotes, this process occurs by the addition of a phosphate group to serine, threonine or tyrosine residues within protein kinases, normally to regulate the catalytic activity.Petsko, GA and Ringe, D 2009, 'Protein Structure and Function', Oxford University Press Inc., New York, U.S.A Autophosphorylation may occur when a kinases' own active site catalyzes the phosphorylation reaction (cis autophosphorylation), or when another kinase of the same type provides the active site that carries out the chemistry (trans autophosphorylation). The latter often occurs when kinase molecules dimerize. In general, the phosphate groups introduced are gamma phosphates from nucleoside triphosphates, most commonly ATP. Function Protein kinases, many of which are regulated by autophosphorylation, are vital in controlling the cellular prolife ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Receptor Tyrosine Kinase
Receptor tyrosine kinases (RTKs) are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinase proteins. Receptor tyrosine kinases have been shown not only to be key regulators of normal cellular processes but also to have a critical role in the development and progression of many types of cancer. Mutations in receptor tyrosine kinases lead to activation of a series of signalling cascades which have numerous effects on protein expression. The receptors are generally activated by dimerization and substrate presentation. Receptor tyrosine kinases are part of the larger family of protein tyrosine kinases, encompassing the receptor tyrosine kinase proteins which contain a transmembrane domain, as well as the non-receptor tyrosine kinases which do not possess transmembrane domains. History The first RTKs to be discovered were the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
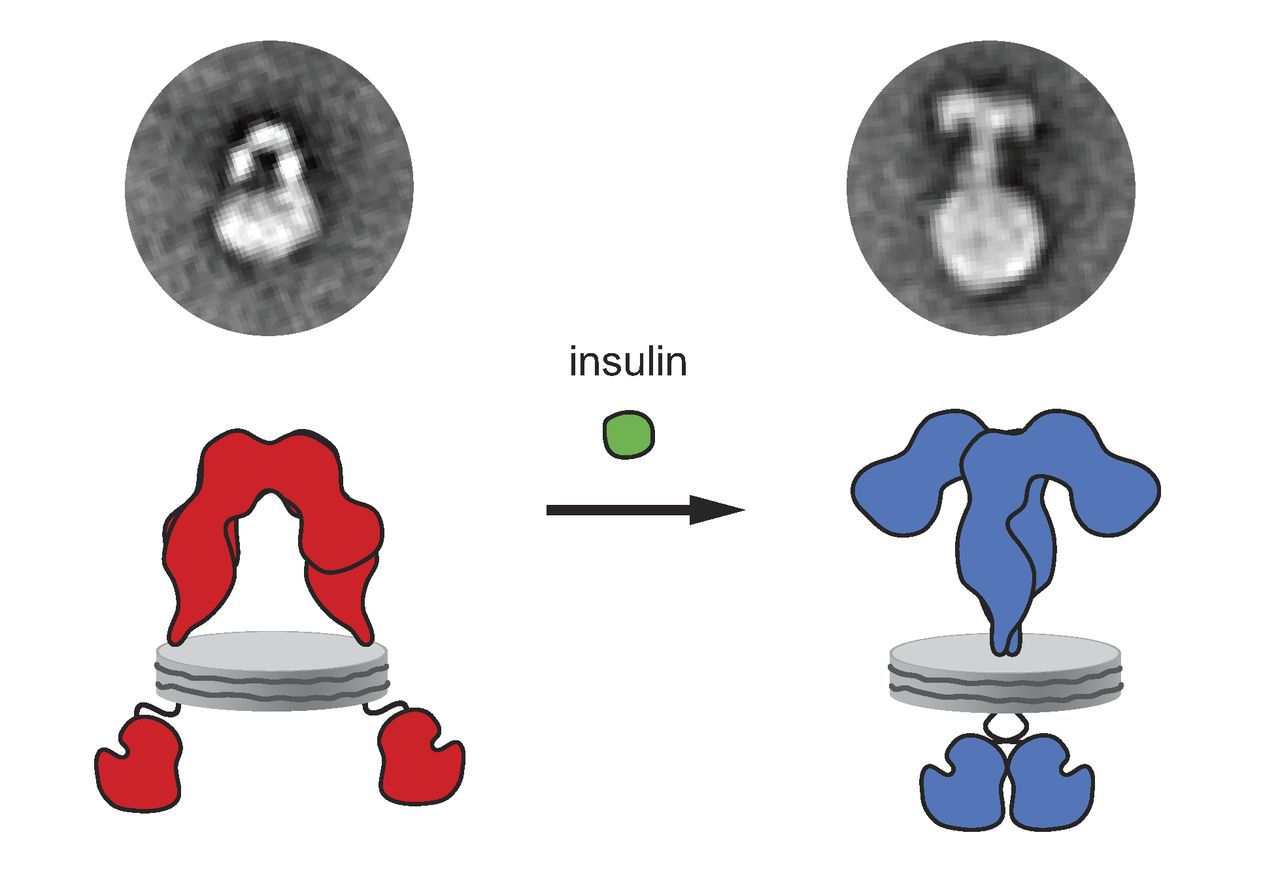
Insulin Receptor
The insulin receptor (IR) is a transmembrane receptor that is activated by insulin, IGF-I, IGF-II and belongs to the large class of receptor tyrosine kinase. Metabolically, the insulin receptor plays a key role in the regulation of glucose homeostasis; a functional process that under degenerate conditions may result in a range of clinical manifestations including diabetes and cancer. Insulin signalling controls access to blood glucose in body cells. When insulin falls, especially in those with high insulin sensitivity, body cells begin only to have access to lipids that do not require transport across the membrane. So, in this way, insulin is the key regulator of fat metabolism as well. Biochemically, the insulin receptor is encoded by a single gene , from which alternate splicing during transcription results in either IR-A or IR-B isoforms. Downstream post-translational events of either isoform result in the formation of a proteolytically cleaved α and β subunit, which up ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the insulin (''INS)'' gene. It is the main Anabolism, anabolic hormone of the body. It regulates the metabolism of carbohydrates, fats, and protein by promoting the absorption of glucose from the blood into cells of the liver, fat cell, fat, and skeletal muscles. In these tissues the absorbed glucose is converted into either glycogen, via glycogenesis, or Fatty acid metabolism#Glycolytic end products are used in the conversion of carbohydrates into fatty acids, fats (triglycerides), via lipogenesis; in the liver, glucose is converted into both. Glucose production and secretion by the liver are strongly inhibited by high concentrations of insulin in the blood. Circulating insulin also affects the synthesis of proteins in a wide variety of tissues. It is thus an anabolic hormone, promoting the conversion of small molecules in the blood into large ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activation Of EGFR By Autophosphorylation
In chemistry and biology, activation is the process whereby something is prepared or excited for a subsequent reaction. Chemistry In chemistry, "activation" refers to the reversible transition of a molecule into a nearly identical chemical or physical state, with the defining characteristic being that this resultant state exhibits an increased propensity to undergo a specified chemical reaction. Thus, activation is conceptually the opposite of protection, in which the resulting state exhibits a ''decreased'' propensity to undergo a certain reaction. The energy of activation specifies the amount of free energy the reactants must possess (in addition to their rest energy) in order to initiate their conversion into corresponding products—that is, in order to reach the transition state for the reaction. The energy needed for activation can be quite small, and often it is provided by the natural random thermal fluctuations of the molecules themselves (i.e. without any externa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen-bonding
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently bonded to a more Electronegativity, electronegative donor atom or group (Dn), interacts with another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Unlike simple Dipole–dipole attraction, dipole–dipole interactions, hydrogen bonding arises from charge transfer (nB → σ*AH), Atomic orbital, orbital interactions, and quantum mechanical Delocalized electron, delocalization, making it a resonance-assisted interaction rather than a mere electrostatic attraction. The general notation for hydrogen bonding is Dn−H···Ac, where the solid line represents a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are nitrogen (N), oxy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
In chemistry, dimerization is the process of joining two identical or similar Molecular entity, molecular entities by Chemical bond, bonds. The resulting bonds can be either strong or weak. Many symmetrical chemical species are described as dimers, even when the monomer is unknown or highly unstable. The term ''homodimer'' is used when the two subunits are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called Dissociation (chemistry), dissociation. When two oppositely-charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Danish chemist Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Many OH-containing molecules form dimers, e.g. the water dimer. Dimers that form based on w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand
In coordination chemistry, a ligand is an ion or molecule with a functional group that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electron pairs, often through Lewis acids and bases, Lewis bases. The nature of metal–ligand bonding can range from covalent bond, covalent to ionic bond, ionic. Furthermore, the metal–ligand bond order can range from one to three. Ligands are viewed as Lewis bases, although rare cases are known to involve Lewis acids and bases, Lewis acidic "ligands". Metals and metalloids are bound to ligands in almost all circumstances, although gaseous "naked" metal ions can be generated in a high vacuum. Ligands in a complex dictate the reactivity (chemistry), reactivity of the central atom, including ligand substitution rates, the reactivity of the ligands themselves, and redox. Ligand selection requires critical consideration in many practical are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signaling Pathway
In biology, cell signaling (cell signalling in British English) is the process by which a cell interacts with itself, other cells, and the environment. Cell signaling is a fundamental property of all cellular life in both prokaryotes and eukaryotes. Typically, the signaling process involves three components: the signal, the receptor, and the effector. In biology, signals are mostly chemical in nature, but can also be physical cues such as pressure, voltage, temperature, or light. Chemical signals are molecules with the ability to bind and activate a specific receptor. These molecules, also referred to as ligands, are chemically diverse, including ions (e.g. Na+, K+, Ca2+, etc.), lipids (e.g. steroid, prostaglandin), peptides (e.g. insulin, ACTH), carbohydrates, glycosylated proteins (proteoglycans), nucleic acids, etc. Peptide and lipid ligands are particularly important, as most hormones belong to these classes of chemicals. Peptides are usually polar, hydrophilic molecules ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activation Loop
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi- domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins. They are most numerous in eukaryotes, with an estimated 30-40% of residues in the eukaryotic proteome located in disordered regions. Disorder is present in around 70% of proteins, either in the form of disordered tails or flexible linkers. Proteins can also be entirely disordered and lack a defined secondary and/or tertiary structure. Their discovery has disproved the idea that three-dimensional ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primary Structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the encoded 22, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically synthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause diseases. Proteolysis can also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |