|
Segmented Filamentous Bacteria
Segmented filamentous bacteria or ''Candidatus'' Savagella are members of the gut microbiota of rodents, fish and chickens, and have been shown to potently induce immune responses in mice. They form a distinct lineage within the Clostridiaceae and the name ''Candidatus'' Savagella has been proposed for this lineage. They were previously named ''Candidatus'' Arthromitus because of their morphological resemblance to bacterial filaments previously observed in the guts of insects by Joseph Leidy. Despite the fact that they have been widely referred to as segmented filamentous bacteria, this term is somewhat problematic as it does not allow one to distinguish between bacteria that colonize various hosts or even if segmented filamentous bacteria are actually several different bacterial species. In mice, these bacteria grow primarily in the terminal ileum in close proximity to the intestinal epithelium where they are thought to help induce T helper 17 cell responses. Intriguingly, S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Candidatus
In prokaryote nomenclature, ''Candidatus'' (Latin for candidate of Roman office) is used to name prokaryotic phyla that are well characterized but yet-uncultured. Contemporary sequencing approaches, such as 16S sequencing or metagenomics, provide much information about the analyzed organisms and thus allow to identify and characterize individual species. However, the majority of prokaryotic species remain uncultivable and hence inaccessible for further characterization in ''in vitro'' study. The recent discoveries of a multitude of candidate taxa has led to candidate phyla radiation expanding the tree of life through the new insights in bacterial diversity. Nomenclature History The initial International Code of Nomenclature of Prokaryotes as well as early revisions did not account for the possibility of identifying prokaryotes which were not yet cultivable. Therefore, the term ''Candidatus'' was proposed in the context of a conference of the International Committee on Systemat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T Helper 17 Cell
T helper 17 cells (Th17) are a subset of pro-inflammatory T helper cells defined by their production of interleukin 17 (IL-17). They are related to T regulatory cells and the signals that cause Th17s to differentiate actually inhibit Treg differentiation. However, Th17s are developmentally distinct from Th1 and Th2 lineages. Th17 cells play an important role in maintaining mucosal barriers and contributing to pathogen clearance at mucosal surfaces; such protective and non-pathogenic Th17 cells have been termed as Treg17 cells. They have also been implicated in autoimmune and inflammatory disorders. The loss of Th17 cell populations at mucosal surfaces has been linked to chronic inflammation and microbial translocation. These regulatory Th17 cells can be generated by TGF-beta plus IL-6 in vitro. Differentiation Like conventional regulatory T cells (Treg), induction of regulatory Treg17 cells could play an important role in modulating and preventing certain autoimmune disease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Google Scholar
Google Scholar is a freely accessible web search engine that indexes the full text or metadata of scholarly literature across an array of publishing formats and disciplines. Released in beta in November 2004, the Google Scholar index includes peer-reviewed online academic journals and books, conference papers, theses and dissertations, preprints, abstracts, technical reports, and other scholarly literature, including court opinions and patents. Google Scholar uses a web crawler, or web robot, to identify files for inclusion in the search results. For content to be indexed in Google Scholar, it must meet certain specified criteria. An earlier statistical estimate published in PLOS One using a mark and recapture method estimated approximately 80–90% coverage of all articles published in English with an estimate of 100 million.''Trend Watch'' (2014) Nature 509(7501), 405 – discussing Madian Khabsa and C Lee Giles (2014''The Number of Scholarly Documents on the Public W ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Bacterial Orders
This article lists the orders of the Bacteria. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) and the phylogeny is based on 16S rRNA-based LTP release 132 by The All-Species Living Tree Project. Phylogeny National Center for Biotechnology Information (NCBI) taxonomy was initially used to decorate the genome tree via tax2tree. The 16S rRNA-based Greengenes taxonomy is used to supplement the taxonomy particularly in regions of the tree with no cultured representatives. List of Prokaryotic names with Standing in Nomenclature (LPSN) is used as the primary taxonomic authority for establishing naming priorities. Taxonomic ranks are normalised using phylorank and the taxonomy manually curated to remove polyphyletic groups. Cladogram was taken from GTDB release 07-RS207 (8th April 2022). Clade Terrabacteria Phylum Chloroflexota * Class ?"Bathosphaeria" Mehrshad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Bacteria Genera
This article lists the genera of the bacteria. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). However many taxonomic names are taken from the GTDB release 07-RS207 (8th April 2022). Phyla {, border="0" style="width: 100%;" ! , - , style="border:0px" valign="top", {, class="wikitable sortable" style="width: 100%; font-size: 95%;" !Syperphylum !Phylum !Authority !Synonyms , - , Parakaryota , , , Myojin parakaryote , - , , " Canglongiota" , Zhang et al. 2022 , , - , , " Fervidibacteria" , , OctSpa1-106 , - , , " Heilongiota" , Zhang et al. 2022 , , - , , " Qinglongiota" , Zhang et al. 2022 , , - , , " Salinosulfoleibacteria" , Tazi et al. 2006 , , - , , " Teskebacteria" , Dojka 1998 , WS1 , - , , " Tharpellota" , Speth et al. 2022 , , - , Terrabacteria , Chloroflexota , Whitman et al. 2018 , " Thermomicrobiota" , - , Terrabacteria , " Dormibacter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mucosal
A mucous membrane or mucosa is a membrane that lines various cavities in the body of an organism and covers the surface of internal organs. It consists of one or more layers of epithelial cells overlying a layer of loose connective tissue. It is mostly of endodermal origin and is continuous with the skin at body openings such as the eyes, eyelids, ears, inside the nose, inside the mouth, lips, the genital areas, the urethral opening and the anus. Some mucous membranes secrete mucus, a thick protective fluid. The function of the membrane is to stop pathogens and dirt from entering the body and to prevent bodily tissues from becoming dehydrated. Structure The mucosa is composed of one or more layers of epithelial cells that secrete mucus, and an underlying lamina propria of loose connective tissue. The type of cells and type of mucus secreted vary from organ to organ and each can differ along a given tract. Mucous membranes line the digestive, respiratory and reproductive tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monocolonization
{{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
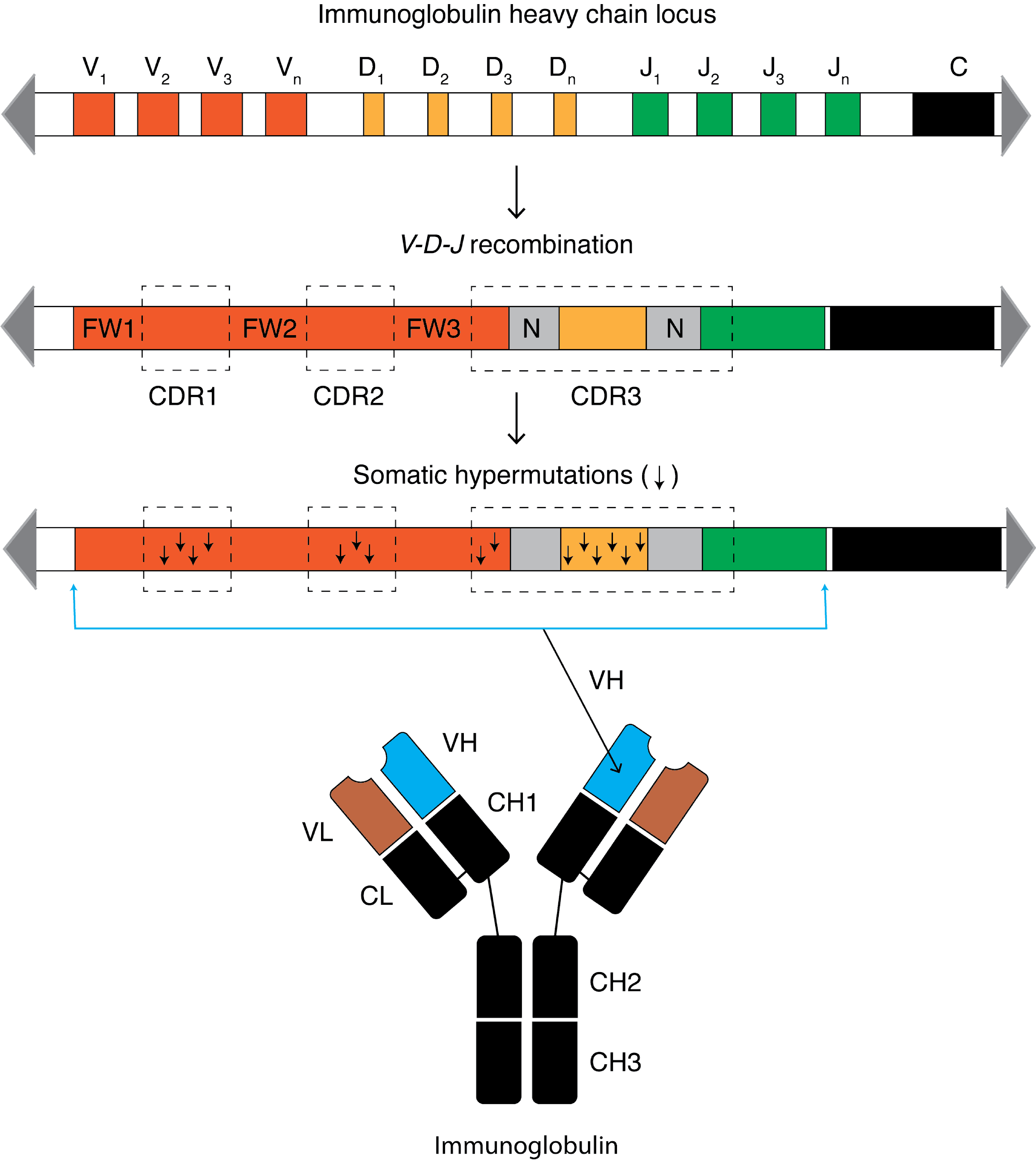
Somatic Hypermutation
Somatic hypermutation (or SHM) is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it (e.g. microbes), as seen during class switching. A major component of the process of affinity maturation, SHM diversifies B cell receptors used to recognize foreign elements (antigens) and allows the immune system to adapt its response to new threats during the lifetime of an organism. Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes. Unlike germline mutation, SHM affects only an organism's individual immune cells, and the mutations are not transmitted to the organism's offspring.Oprea, M. (1999''Antibody Repertoires and Pathogen Recognition:'' The Role of Germline Diversity and Somatic Hypermutation'' (Thesis) University of Leeds.'' Because this mechanism is merely selective and not precisely targeted, somatic hypermutation has been strongly implicated in the development of B-cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Humoral Immune Response
Humoral immunity is the aspect of immunity that is mediated by macromolecules - including secreted antibodies, complement proteins, and certain antimicrobial peptides - located in extracellular fluids. Humoral immunity is named so because it involves substances found in the humors, or body fluids. It contrasts with cell-mediated immunity. Humoral immunity is also referred to as antibody-mediated immunity. The study of the molecular and cellular components that form the immune system, including their function and interaction, is the central science of immunology. The immune system is divided into a more primitive innate immune system and an acquired or adaptive immune system of vertebrates, each of which contain both humoral and cellular immune elements. Humoral immunity refers to antibody production and the coinciding processes that accompany it, including: Th2 activation and cytokine production, germinal center formation and isotype switching, and affinity maturation and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activation-Induced (Cytidine) Deaminase
Activation-induced cytidine deaminase, also known as AICDA, AID and single-stranded DNA cytosine deaminase, is a 24 kDa enzyme which in humans is encoded by the ''AICDA'' gene. It creates mutations in DNA by deamination of cytosine base, which turns it into uracil (which is recognized as a thymine). In other words, it changes a C:G base pair into a U:G mismatch. The cell's DNA replication machinery recognizes the U as a T, and hence C:G is converted to a T:A base pair. During germinal center development of B lymphocytes, AID also generates other types of mutations, such as C:G to A:T. The mechanism by which these other mutations are created is not well understood. It is a member of the APOBEC family. In B cells in the lymph nodes, AID causes mutations that produce antibody diversity, but that same mutation process leads to B cell lymphoma. Function This gene encodes a DNA-editing deaminase that is a member of the cytidine deaminase family. The protein is involved in so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intestinal Epithelium
The intestinal epithelium is the single cell layer that form the Lumen (anatomy), luminal surface (lining) of both the Small intestine, small and large intestine (colon) of the Human gastrointestinal tract, gastrointestinal tract. Composed of simple columnar epithelium, simple columnar epithelial cells, it serves two main functions: absorbing useful substances into the body and restricting the entry of harmful substances. As part of its protective role, the intestinal epithelium forms an important component of the intestinal mucosal barrier. Certain diseases and conditions are caused by functional defects in the intestinal epithelium. On the other hand, various diseases and conditions can lead to its dysfunction which, in turn, can lead to further complications. Structure The intestinal epithelium is part of the Gastrointestinal tract#mucosa, intestinal mucosa. The epithelium is composed of a single layer of cells, while the other two layers of the mucosa, the lamina propria a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gut Microbiota
Gut microbiota, gut microbiome, or gut flora, are the microorganisms, including bacteria, archaea, fungi, and viruses that live in the digestive tracts of animals. The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota. The gut is the main location of the human microbiome. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis. The microbial composition of the gut microbiota varies across regions of the digestive tract. The colon contains the highest microbial density recorded in any habitat on Earth, representing between 300 and 1000 different species. Bacteria are the largest and to date, best studied component and 99% of gut bacteria come from about 30 or 40 species. Up to 60% of the dry mass of feces is bacteria. Over 99% of the bact ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |