|
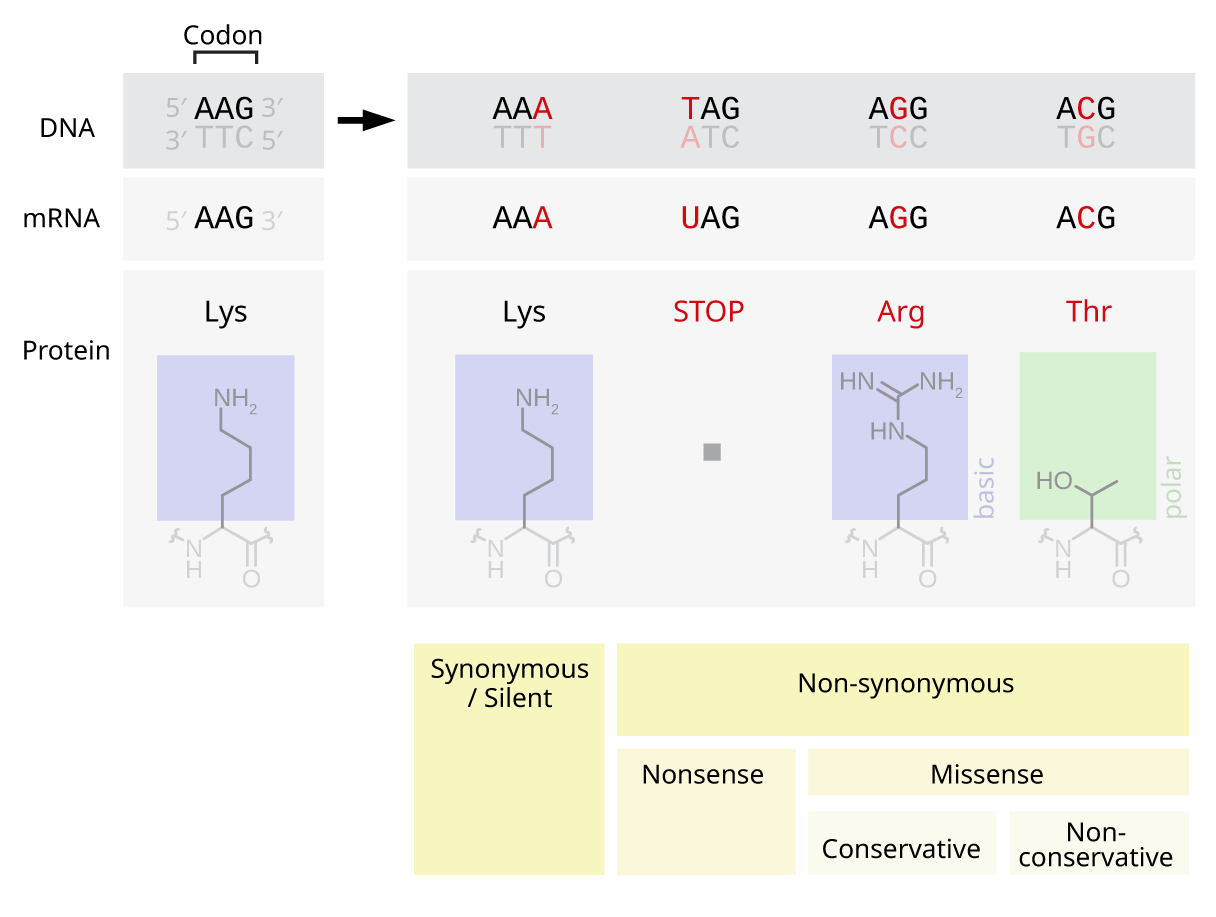
Substitution Matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a Nucleic acid sequence, nucleotide sequence or a Protein primary structure, protein sequence changes to other character states over evolutionary time. The information is often in the form of Logit, log odds of finding two specific character states aligned and depends on the assumed number of evolutionary changes or sequence dissimilarity between compared sequences. It is an application of a stochastic matrix. Substitution matrices are usually seen in the context of amino acid or DNA sequence alignments, where they are used to calculate similarity scores between the aligned sequences. Background In the process of evolution, from one generation to the next the amino acid sequences of an organism's proteins are gradually altered through the action of DNA mutations. For example, the sequence ALEIRYLRD could mutate into the sequence ALEINYLRD in one step, and pos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, data science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The process of analyzing and interpreting data can sometimes be referred to as computational biology, however this distinction between the two terms is often disputed. To some, the term ''computational biology'' refers to building and using models of biological systems. Computational, statistical, and computer programming techniques have been used for In silico, computer simulation analyses of biological queries. They include reused specific analysis "pipelines", particularly in the field of genomics, such as by the identification of genes and single nucleotide polymorphis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein, after Protein biosynthesis, synthesis by a ribosome as a linear chain of Amino acid, amino acids, changes from an unstable random coil into a more ordered protein tertiary structure, three-dimensional structure. This structure permits the protein to become biologically functional or active. The folding of many proteins begins even during the translation of the polypeptide chain. The amino acids interact with each other to produce a well-defined three-dimensional structure, known as the protein's native state. This structure is determined by the amino-acid sequence or primary structure. The correct three-dimensional structure is essential to function, although some parts of functional proteins Intrinsically unstructured proteins, may remain unfolded, indicating that protein dynamics are important. Failure to fold into a native structure generally produces inactive proteins, but in some instances, misfolded proteins have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neighbor-joining
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees, created by Naruya Saitou and Masatoshi Nei in 1987. Usually based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) to create the phylogenetic tree. The algorithm Neighbor joining takes a distance matrix, which specifies the distance between each pair of taxa, as input. The algorithm starts with a completely unresolved tree, whose topology corresponds to that of a star network, and iterates over the following steps, until the tree is completely resolved, and all branch lengths are known: # Based on the current distance matrix, calculate a matrix Q (defined below). # Find the pair of distinct taxa i and j (i.e. with i \neq j) for which Q(i,j) is smallest. Make a new node that joins the taxa i and j, and connect the new node to the central node. For example, in part (B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maximum Likelihood
In statistics, maximum likelihood estimation (MLE) is a method of estimating the parameters of an assumed probability distribution, given some observed data. This is achieved by maximizing a likelihood function so that, under the assumed statistical model, the observed data is most probable. The point in the parameter space that maximizes the likelihood function is called the maximum likelihood estimate. The logic of maximum likelihood is both intuitive and flexible, and as such the method has become a dominant means of statistical inference. If the likelihood function is differentiable, the derivative test for finding maxima can be applied. In some cases, the first-order conditions of the likelihood function can be solved analytically; for instance, the ordinary least squares estimator for a linear regression model maximizes the likelihood when the random errors are assumed to have normal distributions with the same variance. From the perspective of Bayesian inference, ML ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST (biotechnology)
In bioinformatics, BLAST (basic local alignment search tool) is an algorithm and program for comparing Primary structure, primary biological sequence information, such as the amino acid, amino-acid sequences of proteins or the nucleotides of DNA sequence, DNA and/or RNA sequences. A BLAST search enables a researcher to compare a subject protein or nucleotide sequence (called a query) with a library or database of sequences, and identify database sequences that resemble the query sequence above a certain threshold. For example, following the discovery of a previously unknown gene in the Mus musculus, mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. Background BLAST is one of the most widely used bioinformatics programs for sequence searching. It addresses a fundamental problem in bioinformatics research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Steven Henikoff
Steven Henikoff is a scientist at the Fred Hutchinson Cancer Research Center, and an HHMI Investigator. His field of study is chromatin-related transcriptional regulation. He earned his BS in chemistry at the University of Chicago. He earned his PhD in biochemistry and molecular biology from Harvard University in the lab of Matt Meselson in 1977. He did a postdoctoral fellowship at the University of Washington. His research has been funded by the National Science Foundation, National Institutes of Health, and HHMI. In 1992, Steven Henikoff, together with his wife Jorja Henikoff, introduced the BLOSUM substitution matrices. The BLOSUM matrices are widely used for sequence alignment of proteins. In 2005, Henikoff was elected to the National Academy of Sciences The National Academy of Sciences (NAS) is a United States nonprofit, NGO, non-governmental organization. NAS is part of the National Academies of Sciences, Engineering, and Medicine, along with the National Academy of Eng ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLOSUM
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based on local alignments. BLOSUM matrices were first introduced in a paper by Steven Henikoff and Jorja Henikoff. They scanned the BLOCKS database for very conserved regions of protein families (that do not have gaps in the sequence alignment) and then counted the relative frequencies of amino acids and their substitution probabilities. Then, they calculated a log-odds score for each of the 210 possible substitution pairs of the 20 standard amino acids. All BLOSUM matrices are based on observed alignments; they are not extrapolated from comparisons of closely related proteins like the PAM Matrices. Biological background The genetic instructions of every replicating cell in a living organism are contained within its DNA. Throughout the cell's ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Margaret Oakley Dayhoff
Margaret Belle (Oakley) Dayhoff (March 11, 1925 – February 5, 1983) was an American Biophysicist and a pioneer in the field of bioinformatics. Dayhoff was a professor at Georgetown University Medical Center and a noted research biochemist at the National Biomedical Research Foundation, where she pioneered the application of mathematics and computational methods to the field of biochemistry. She dedicated her career to applying the evolving computational technologies to support advances in biology and medicine, most notably the creation of protein and nucleic acid databases and tools to interrogate the databases. She originated one of the first substitution matrices, point accepted mutations (''PAM''). The one-letter code used for amino acids was developed by her, reflecting an attempt to reduce the size of the data files used to describe amino acid sequences in an era of punch-card computing. Her PhD degree was from Columbia University in the department of chemistr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point Accepted Mutation
A point accepted mutation — also known as a PAM — is the replacement of a single amino acid in the primary structure of a protein with another single amino acid, which is accepted by the processes of natural selection. This definition does not include all point mutations in the DNA of an organism. In particular, silent mutations are not point accepted mutations, nor are mutations that are lethal or that are rejected by natural selection in other ways. A PAM matrix is a matrix where each column and row represents one of the twenty standard amino acids. In bioinformatics, PAM matrices are sometimes used as substitution matrices to score sequence alignments for proteins. Each entry in a PAM matrix indicates the likelihood of the amino acid of that row being replaced with the amino acid of that column through a series of one or more point accepted mutations during a specified evolutionary interval, rather than these two amino acids being aligned due to chance. Different PAM matri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Score (statistics)
In statistics, the score (or informant) is the gradient of the log-likelihood function with respect to the statistical parameter, parameter vector. Evaluated at a particular value of the parameter vector, the score indicates the steepness of the log-likelihood function and thereby the sensitivity to infinitesimal changes to the parameter values. If the log-likelihood function is Continuous function, continuous over the parameter space, the score will vanish (mathematics), vanish at a local Maxima and minima, maximum or minimum; this fact is used in maximum likelihood estimation to find the parameter values that maximize the likelihood function. Since the score is a function of the Realization (probability), observations, which are subject to sampling error, it lends itself to a test statistic known as ''score test'' in which the parameter is held at a particular value. Further, the likelihood ratio, ratio of two likelihood functions evaluated at two distinct parameter values can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Log-odds
In statistics, the logit ( ) function is the quantile function associated with the standard logistic distribution. It has many uses in data analysis and machine learning, especially in data transformations. Mathematically, the logit is the inverse of the standard logistic function \sigma(x) = 1/(1+e^), so the logit is defined as : \operatorname p = \sigma^(p) = \ln \frac \quad \text \quad p \in (0,1). Because of this, the logit is also called the log-odds since it is equal to the logarithm of the odds \frac where is a probability. Thus, the logit is a type of function that maps probability values from (0, 1) to real numbers in (-\infty, +\infty), akin to the probit function. Definition If is a probability, then is the corresponding odds; the of the probability is the logarithm of the odds, i.e.: : \operatorname(p)=\ln\left( \frac \right) =\ln(p)-\ln(1-p)=-\ln\left( \frac-1\right)=2\operatorname(2p-1). The base of the logarithm function used is of little importance ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |