|
Ribosome-binding Site
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs. Prokaryotes The RBS in prokaryotes is a region upstream of the start codon. This region of the mRNA has the consensus 5'-AGGAGG-3', also called the Shine-Dalgarno (SD) sequence. The complementary sequence (CCUCCU), called the anti-Shine-Dalgarno (ASD) is contained in the 3’ end of the 16S region of the smaller (30S) ribosomal subunit. Upon encountering the Shine-Dalgarno sequence, the ASD of the ribosome base pairs with it, after which translation is initiated. Variations of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Start Codon
The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and Archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids. The most common start codon is AUG (i.e., ATG in the corresponding DNA sequence). The start codon is often preceded by a 5' untranslated region ( 5' UTR). In prokaryotes this includes the ribosome binding site. Alternative start codons Alternative start codons are different from the standard AUG codon and are found in both prokaryotes (bacteria and archaea) and eukaryotes. Alternate start codons are still translated as Met when they are at the start of a protein (even if the codon encodes a different amino acid otherwise). This is because a separate transfer RNA (tRNA) is used for initiation. Eukaryotes Alternate start codons (non-AUG) are very rare in eukaryotic genomes. However, naturally occurring non-AUG start codons have been rep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Artificial Neural Network
Artificial neural networks (ANNs), usually simply called neural networks (NNs) or neural nets, are computing systems inspired by the biological neural networks that constitute animal brains. An ANN is based on a collection of connected units or nodes called artificial neurons, which loosely model the neurons in a biological brain. Each connection, like the synapses in a biological brain, can transmit a signal to other neurons. An artificial neuron receives signals then processes them and can signal neurons connected to it. The "signal" at a connection is a real number, and the output of each neuron is computed by some non-linear function of the sum of its inputs. The connections are called ''edges''. Neurons and edges typically have a ''weight'' that adjusts as learning proceeds. The weight increases or decreases the strength of the signal at a connection. Neurons may have a threshold such that a signal is sent only if the aggregate signal crosses that threshold. Typically ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Biosynthesis
Protein biosynthesis (or protein synthesis) is a core biological process, occurring inside cells, balancing the loss of cellular proteins (via degradation or export) through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences. Protein synthesis can be divided broadly into two phases - transcription and translation. During transcription, a section of DNA encoding a protein, known as a gene, is converted into a template molecule called messenger RNA (mRNA). This conversion is carried out by enzymes, known as RNA polymerases, in the nucleus of the cell. In eukaryotes, this mRNA is initially produced in a premature form (pre-mRNA) which undergoes post-transcriptional modifications to produce mature mRNA. The mature mRNA is exported from the cell nucleus via nuclear pores to the cytoplasm of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Prediction
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions. Gene finding is one of the first and most important steps in understanding the genome of a species once it has been sequenced. In its earliest days, "gene finding" was based on painstaking experimentation on living cells and organisms. Statistical analysis of the rates of homologous recombination of several different genes could determine their order on a certain chromosome, and information from many such experiments could be combined to create a genetic map specifying the rough location of known genes relative to each other. Today, with comprehensive genome sequence and powerful computational resources at the disposal of the research community, gene finding has been redefined as a largely computatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaeal Translation
Archaeal translation is the process by which messenger RNA is translated into proteins in archaea. Not much is known on this subject, but on the protein level it seems to resemble eukaryotic translation. Most of the initiation, elongation, and termination factors in archaea have homologs in eukaryotes. Shine-Dalgarno sequences only are found in a minority of genes for many phyla, with many leaderless mRNAs probably initiated by scanning. The process of ABCE1 ATPase-based recycling is also shared with eukaryotes. Being a prokaryote A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ... without a nucleus, archaea do perform transcription and translation at the same time like bacteria do. References Molecular biology Protein biosynthesis Gene expression {{Molecular-biology-st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Translation
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria. Initiation Initiation of translation in bacteria involves the assembly of the components of the translation system, which are: the two ribosomal subunits ( 50S and 30S subunits); the mature mRNA to be translated; the tRNA charged with N-formylmethionine (the first amino acid in the nascent peptide); guanosine triphosphate (GTP) as a source of energy, and the three prokaryotic initiation factors IF1, IF2, and IF3, which help the assembly of the initiation complex. Variations in the mechanism can be anticipated. The ribosome has three active sites: the A site, the P site, and the E site. The ''A site'' is the point of entry for the aminoacyl tRNA (except for the first aminoacyl tRNA, which enters at the P site). The ''P site'' is where the peptidyl tRNA is formed in the ribosome. And the ''E site'' which is the exit site of the now uncharged tRNA after it gives its amino acid to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Translation
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: gene translation, elongation, termination, and recapping. Initiation Translation initiation is the process by which the ribosome and its associated factors bind to an mRNA and are assembled at the start codon. This process is defined as either cap-dependent, in which the ribosome binds initially at the 5' cap and then travels to the stop codon, or as cap-independent, where the ribosome does not initially bind the 5' cap. Cap-dependent initiation Initiation of translation usually involves the interaction of certain key proteins, the initiation factors, with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap, as well as with the 5' UTR. These proteins bind the small (40S) ribosomal subunit and hold the mRNA in place. eIF3 is associated with the 40S ribosomal subunit and plays a role in keeping the large (60S) ribosomal sub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Operon Ribosome Binding Site
The alpha operon ribosome binding site in bacteria is surrounded by this complex pseudoknotted RNA structure. Translation of the mRNA produces 4 ribosomal protein products, one of which (S4) acts as a translational repressor by binding to the nested pseudoknot region. The mechanism of repression is thought to involve a conformational switch in the pseudoknot region and ribosome Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to ... entrapment. References External links * Cis-regulatory RNA elements {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
University Of Pittsburgh
The University of Pittsburgh (Pitt) is a public state-related research university in Pittsburgh, Pennsylvania. The university is composed of 17 undergraduate and graduate schools and colleges at its urban Pittsburgh campus, home to the university's central administration and around 28,000 undergraduate and graduate students. The 132-acre Pittsburgh campus includes various historic buildings that are part of the Schenley Farms Historic District, most notably its 42-story Gothic revival centerpiece, the Cathedral of Learning. Pitt is a member of the Association of American Universities and is classified among "R1: Doctoral Universities – Very high research activity". It is the second-largest non-government employer in the Pittsburgh metropolitan area. Pitt traces its roots to the Pittsburgh Academy founded by Hugh Henry Brackenridge in 1787. While the city was still on the edge of the American frontier at the time, Pittsburgh's rapid growth meant that a proper university was so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Marilyn Kozak
Marilyn S. Kozak is an American professor of biochemistry at the Robert Wood Johnson Medical School. She was previously at the University of Medicine and Dentistry of New Jersey before the school was merged. She was awarded a PhD in microbiology by Johns Hopkins University studying the synthesis of the Bacteriophage MS2, advised by Daniel Nathans. In her original faculty job proposal, she sought to study the mechanism of eukaryotic translation initiation, a problem long thought to have already been solved by Joan Steitz. While in the Department of Biological Sciences at University of Pittsburgh, she published a series of studies that established the scanning model of translation initiation and the Kozak consensus sequence. Her current research interests are unknown as her last publication was in 2008. Recognition Marilyn Kozak was listed as one of the top 10 Women Scientists of the 80's in an article published by The Scientist. This was awarded based on the number of citation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
John Shine
John Shine (born 3 July 1946) is an Australian biochemist and molecular biologist. Shine and Lynn Dalgarno discovered the nucleotide sequence, called the Shine-Dalgarno sequence, necessary for the initiation and termination of protein synthesis. He directed the Garvan Institute of Medical Research in Sydney from 1990 to 2011. In May 2018 Shine was elected President of the Australian Academy of Science. Background and early career The brother of scientist, Richard Shine, John Shine was born in Brisbane in 1946 and completed his university studies at the Australian National University (ANU) in Canberra, graduating with a bachelor of science with honours in 1972 and completing his PhD in 1975. During the course of his studies he and his supervisor, Lynn Dalgarno, discovered the RNA sequence necessary for ribosome binding and the initiation of protein synthesis in the bacterium ''Escherichia coli''. The sequence was named the Shine-Dalgarno sequence. This was a key discover ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
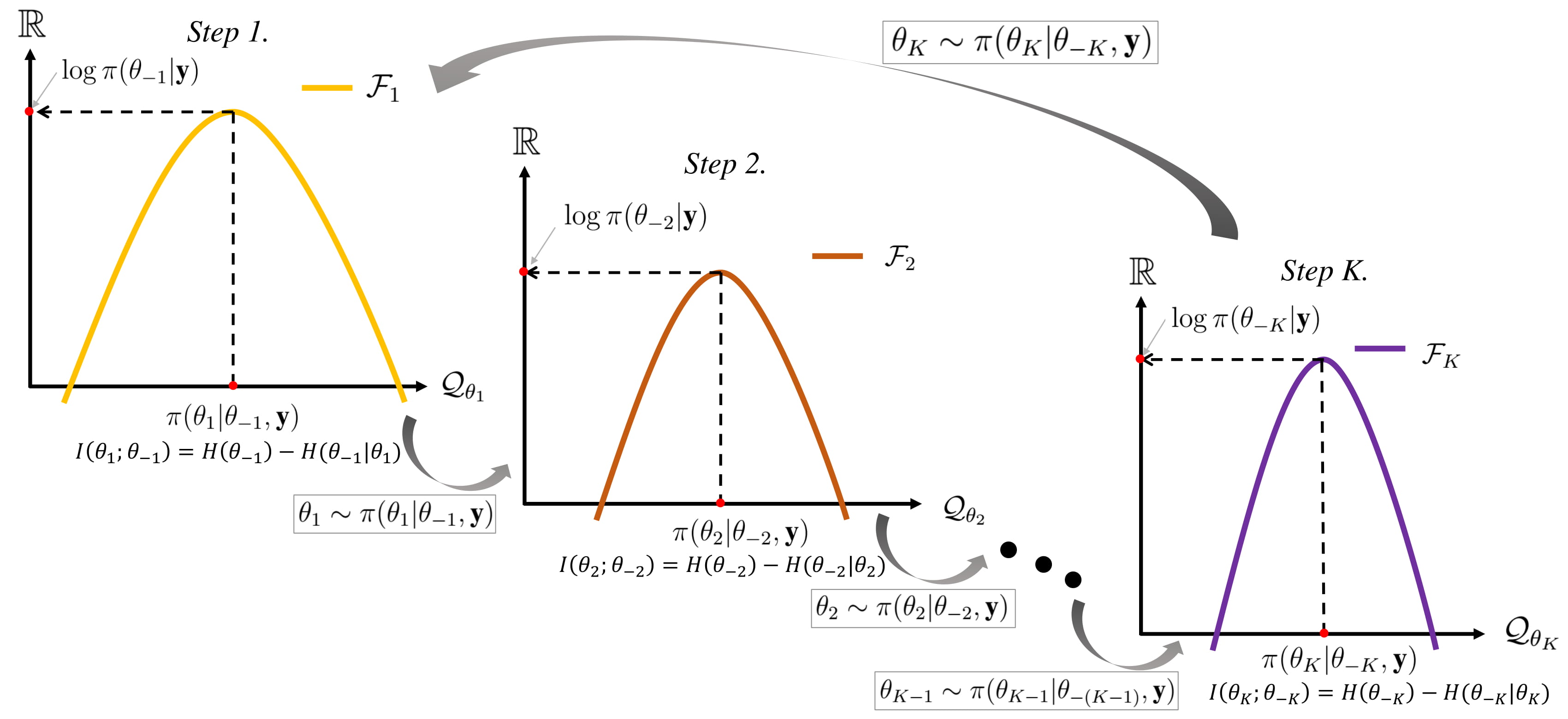
Gibbs Sampling
In statistics, Gibbs sampling or a Gibbs sampler is a Markov chain Monte Carlo (MCMC) algorithm for obtaining a sequence of observations which are approximated from a specified multivariate probability distribution, when direct sampling is difficult. This sequence can be used to approximate the joint distribution (e.g., to generate a histogram of the distribution); to approximate the marginal distribution of one of the variables, or some subset of the variables (for example, the unknown parameters or latent variables); or to compute an integral (such as the expected value of one of the variables). Typically, some of the variables correspond to observations whose values are known, and hence do not need to be sampled. Gibbs sampling is commonly used as a means of statistical inference, especially Bayesian inference. It is a randomized algorithm (i.e. an algorithm that makes use of random numbers), and is an alternative to deterministic algorithms for statistical inferenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |