|
RasMol
RasMol is a computer program written for molecular graphics visualization intended and used mainly to depict and explore biological macromolecule structures, such as those found in the Protein Data Bank (PDB). History It was originally developed by Roger Sayle in the early 1990s. Historically, it was an important tool for molecular biologists since the extremely optimized program allowed the software to run on (then) modestly powerful personal computers. Before RasMol, visualization software ran on graphics workstations that, due to their cost, were less accessible to scholars. RasMol continues to be important for research in structural biology, and has become important in education. RasMol has a complex licensing version history. Starting with the version 2.7 series, RasMol source code is dual-licensed under a GNU General Public License (GPL), or custom license ''RASLIC''. Starting with version 2.7.5, a GPL is the only license valid for binary distributions. RasMol include ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sirius Visualization Software
Sirius is a molecular modelling and analysis system developed at San Diego Supercomputer Center. Sirius is designed to support advanced user requirements that go beyond simple display of small molecules and proteins. Sirius supports high quality interactive 3D graphics, structure building, displaying protein or DNA primary sequences, access to remote data sources, and visualizing molecular dynamics trajectories. It can be used for scientific visualization and analysis, and chemistry and biology instruction. This software is no longer supported as of 2011. Key features Sirius supports a variety of applications with a set of features, including: * Building and editing chemical structures using a library of fragments * Protein structure and sequence alignment * Command line interpreter and scripting support fully compatible with extant RasMol scripts * Full support for molecular dynamics trajectory visualizing * BLAST search directly in Protein Data Bank and Uniprot databases * A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Molecular Graphics Systems
This is a list of notable software systems that are used for visualizing macromolecules. Key The tables below indicate which types of data can be visualized in each system: See also * Biological data visualization * Comparison of nucleic acid simulation software * Comparison of software for molecular mechanics modeling * List of microscopy visualization systems * List of open-source bioinformatics software * Molecular graphics * Molecule editor A notable molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional ... References External links * A rather detailed, objective, and technical assessment of about 20 tools. * * * {{Chemistry software Chemistry software molecular graphics systems Molecular modelling Computational chemistry software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Free And Open-source Software Packages
This is a list of free and open-source software (FOSS) packages, computer software licensed under free software licenses and open-source licenses. Software that fits the Free Software Definition may be more appropriately called free software; the GNU project in particular objects to their works being referred to as open-source. For more information about the philosophical background for open-source software, see free software movement and Open Source Initiative. However, nearly all software meeting the Free Software Definition also meets '' the Open Source Definition'' and vice versa. A small fraction of the software that meets either definition is listed here. Some of the open-source applications are also the basis of commercial products, shown in the List of commercial open-source applications and services. Artificial intelligence General AI * OpenCog – A project that aims to build an artificial general intelligence (AGI) framework. OpenCog Prime is a specific set ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
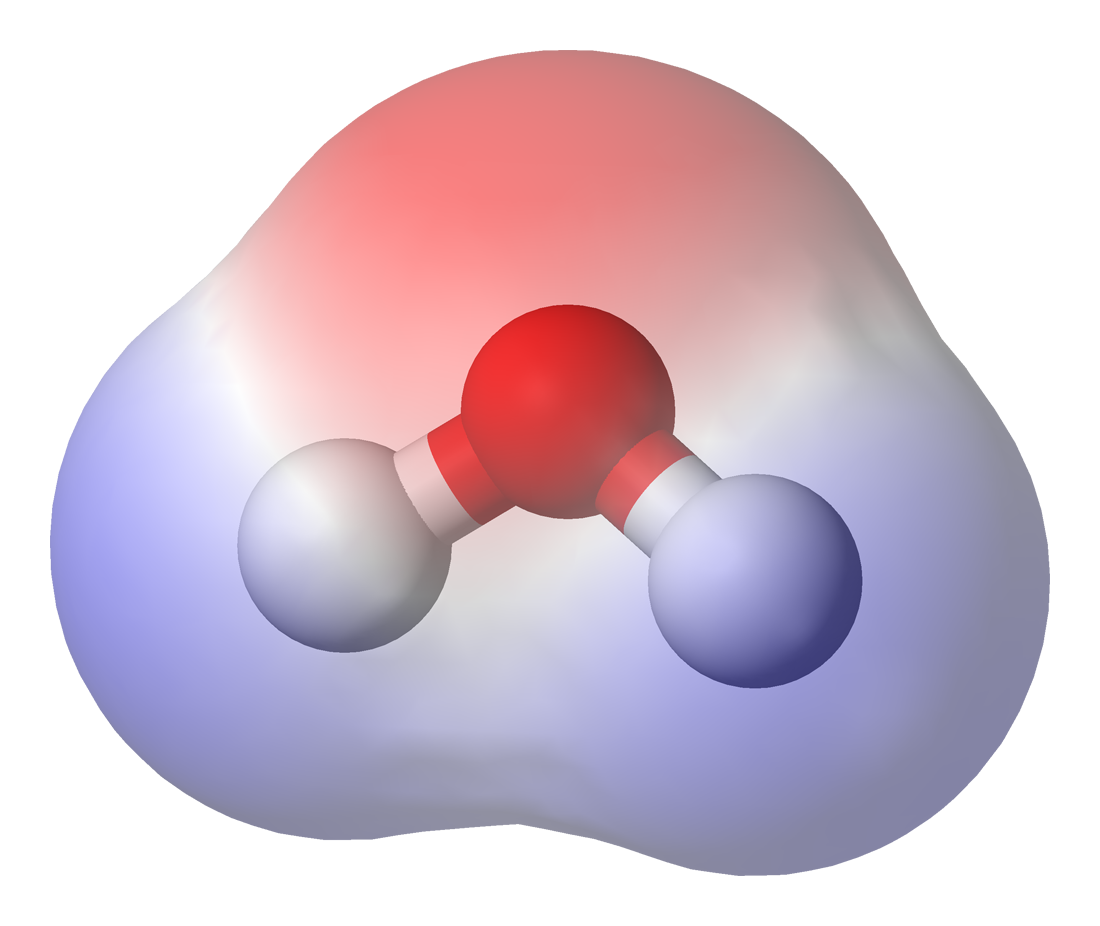
Molecular Graphics
Molecular graphics is the discipline and philosophy of studying molecules and their properties through graphical representation. IUPAC limits the definition to representations on a "graphical display device". Ever since Dalton's atoms and Kekulé's benzene, there has been a rich history of hand-drawn atoms and molecules, and these representations have had an important influence on modern molecular graphics. Colour molecular graphics are often used on chemistry journal covers artistically. History Prior to the use of computer graphics in representing molecular structure, Robert Corey and Linus Pauling developed a system for representing atoms or groups of atoms from hard wood on a scale of 1 inch = 1 angstrom connected by a clamping device to maintain the molecular configuration. These early models also established the CPK coloring scheme that is still used today to differentiate the different types of atoms in molecular models (e.g. carbon = black, oxygen = red, nitrogen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Graphics
Molecular graphics is the discipline and philosophy of studying molecules and their properties through graphical representation. IUPAC limits the definition to representations on a "graphical display device". Ever since Dalton's atoms and Kekulé's benzene, there has been a rich history of hand-drawn atoms and molecules, and these representations have had an important influence on modern molecular graphics. Colour molecular graphics are often used on chemistry journal covers artistically. History Prior to the use of computer graphics in representing molecular structure, Robert Corey and Linus Pauling developed a system for representing atoms or groups of atoms from hard wood on a scale of 1 inch = 1 angstrom connected by a clamping device to maintain the molecular configuration. These early models also established the CPK coloring scheme that is still used today to differentiate the different types of atoms in molecular models (e.g. carbon = black, oxygen = red, nitrogen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank (file Format)
The Protein Data Bank (PDB) file format is a textual file format describing the three-dimensional structures of molecules held in the Protein Data Bank, now succeeded by the Macromolecular Crystallographic Information File, mmCIF format. The PDB format accordingly provides for description and annotation of protein and nucleic acid structures including atomic coordinates, secondary structure assignments, as well as atomic connectivity. In addition experimental metadata are stored. The PDB format is the legacy file format for the Protein Data Bank which has kept data on biological macromolecules in the newer Macromolecular Crystallographic Information File, PDBx/mmCIF file format since 2014. History The PDB file format was invented in 1972 as a human-readable file that would allow researchers to exchange the atomic coordinates in a given protein through a database system. Its fixed-column width format is limited to 80 or 140 columns, which was based on the width of the computer punch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiple Sequence Alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis and can highlight homologous features between sequences. Alignments highlight mutation events such as point mutations (single amino acid or nucleotide changes), insertion mutations and deletion mutations, and alignments are used to assess sequence conservation and infer the presence and activity of protein domains, tertiary structures, secondary structures, and individual amino acids or nucleotides. Multiple sequence alignments require more sophisticated methodologies than pairwise alignments, as they are more computationally complex. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibiti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dynamic Data Exchange
In computing, Dynamic Data Exchange (DDE) is a technology for interprocess communication used in early versions of Microsoft Windows and OS/2. DDE allows programs to manipulate objects provided by other programs, and respond to user actions affecting those objects. DDE was partially superseded by Object Linking and Embedding (OLE), and is currently maintained in Windows systems only for the sake of backward compatibility. History and architecture Dynamic Data Exchange was first introduced in 1987 with the release of Windows 2.0 as a method of interprocess communication so that one program could communicate with or control another program, somewhat like Sun's RPC (Remote Procedure Call). At the time, the only method for communication between the operating system and client applications was the "Windows Messaging Layer." DDE extended this protocol to allow peer-to-peer communication among client applications, via message broadcasts. Because DDE runs via message broadcasts, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also *Car–Parrinello molecular dynamics *Comparison of force-field implementations *Comparison of nucleic acid simulation software *List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling *List of software for nanostructures modeling *Molecular design software *Molecular dynamics *Molecular modeling on GPUs *Molecule editor *PyMOL Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tk (software)
Tk is a cross-platform widget toolkit that provides a library of basic elements of GUI widgets for building a graphical user interface (GUI) in many programming languages. It is free and open-source software released under a BSD-style software license. Tk provides many widgets commonly needed to develop desktop applications, such as button, menu, canvas, text, frame, label, etc. Tk has been ported to run on most flavors of Linux, macOS, Unix, and Microsoft Windows. Like Tcl, Tk supports Unicode within the Basic Multilingual Plane, but it has not yet been extended to handle the current extended full Unicode (e.g., UTF-16 from UCS-2 that Tk supports). Tk was designed to be extended, and a wide range of extensions are available that offer new widgets or other capabilities. Since Tcl/Tk 8, it offers "native look and feel" (for instance, menus and buttons are displayed in the manner of "native" software for any given platform). Highlights of version 8.5 include a new theming engine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecule Editor
A notable molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, via 2D computer graphics or 3D computer graphics, respectively. Two-dimensional output is used as illustrations or to query chemical databases. Three-dimensional output is used to build molecular models, usually as part of molecular modelling software packages. Database molecular editors such as Leatherface, RECAP, and Molecule Slicer allow large numbers of molecules to be modified automatically according to rules such as 'deprotonate carboxylic acids' or 'break exocyclic bonds' that can be specified by a user. Molecule editors typically support reading and writing at least one file format or line notation. Examples of each include Molfile and simplified molecular input line entry specification (SMILES), respectively. Files ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |