|
Nucleic Acid Analogues
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix). Nucleic acid analogues are also called xeno nucleic acids and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries. Artificial nucleic acids include peptide nucleic acids (PNA), morpholino, and locked nu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Difference DNA RNA-EN
Difference commonly refers to: * Difference (philosophy), the set of properties by which items are distinguished * Difference (mathematics), the result of a subtraction Difference, The Difference, Differences or Differently may also refer to: Music * Difference (album), ''Difference'' (album), by Dreamtale, 2005 * The Difference (album), ''The Difference'' (album), Pendleton, 2008 * The Difference (The Wallflowers song), "The Difference" (The Wallflowers song), 1997 * Differences (song), "Differences" (song), by Ginuwine, 2001 * Differently (album), ''Differently'' (album), by Cassie Davis, 2009 ** Differently (song), "Differently" (song), by Cassie Davis, 2009 * "Difference", a song by Benjamin Clementine from the 2022 album ''And I Have Been'' * "The Difference", a song by Matchbox Twenty from the 2002 album ''More Than You Think You Are'' * "The Difference", a song by Westlife from the 2009 album ''Where We Are'' * "The Difference", a song by Nick Jonas from the 2016 album ''L ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyelectrolyte Theory Of The Gene
The polyelectrolyte theory of the gene proposes that for a linear genetic biopolymer dissolved in water, such as DNA, to undergo Evolution, Darwinian evolution anywhere in the universe, it must be a polyelectrolyte, a polymer containing repeating Ionic charge, ionic charges. These charges maintain the uniform physical properties needed for Darwinian evolution, regardless of the information encoded in the Genetics, genetic biopolymer. DNA is such a molecule. Regardless of its nucleic acid sequence, the negative charges on its backbone dominate the physical interactions of the molecule to such a degree that it maintains uniform physical properties such as its Aqueous solution, aqueous solubility and double-helix structure. The polyelectrolyte theory of the gene was proposed by Steven A. Benner and Daniel Hutter in 2002 and has largely remained a theoretical framework astrobiologists have used to think about how life may be detected beyond Earth. This idea was later linked by Benner to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide Synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides ( dA, dC, dG, and T), ribonucleosides ( A, C, G, and U), or chemically modified nucleosides, e.g. LNA or BNA. To obtain the desired oligonucleotide, the building blocks are sequentially coupled to the growing oligonucleotide chain in the order required by the sequence of the product (s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Nucleic Acid
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA. Synthetic peptide nucleic acid oligomers have been used in recent years in molecular biology procedures, diagnostic assays, and antisense therapies. Due to their higher binding strength, it is not necessary to design long PNA oligomers for use in these roles, which usually require oligonucleotide probes of 20–25 bases. The main concern of the length of the PNA-oligomers is to guarantee the specificity. PNA oligomers also show greater specificity in binding to complementary DNAs, with a PNA/DNA base mismatch being more destabilizing than a similar mismatch in a DNA/DNA duplex. This binding strength and specificity also applies to PNA/RNA duplexes. PNAs are not easily recognized by either nucleases or proteases, making them resistant to degradation by enzymes. PNAs are also stable over a wide pH range. Though an unmodified PNA cannot readily cross the cell membrane to enter the cytosol, co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
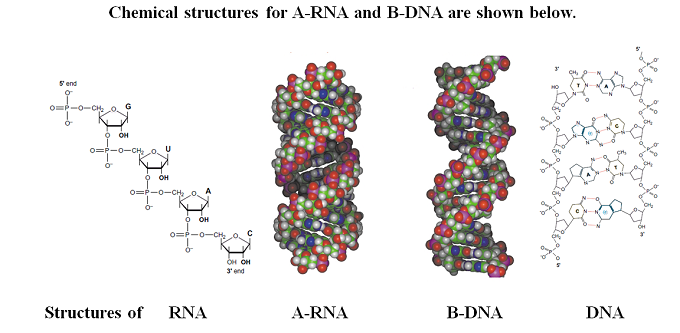
Bridged Nucleic Acid
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering. The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoramidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability. Chemical structures Chemical structures of BNA monomers containing a bridge at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer as synthesized by Takeshi Imanishi's group. The nature of the bridge can vary for different types of monomers. The 3D structures for A-RNA and B-DNA were used as a template for the design of the BNA monomers. The goal for the design was to find deriv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally occurring form, , is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared . Like most sugars, ribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Hydrolysis
RNA hydrolysis is a reaction in which a phosphodiester bond in the sugar-phosphate backbone of RNA is broken, cleaving the RNA molecule. RNA is susceptible to this base-catalyzed hydrolysis because the ribose sugar in RNA has a hydroxyl group at the 2’ position. This feature makes RNA chemically unstable compared to DNA, which does not have this 2’ -OH group and thus is not susceptible to base-catalyzed hydrolysis. Mechanism RNA hydrolysis occurs when the Deprotonation, deprotonated 2’ OH of the ribose, acting as a nucleophile, attacks the adjacent phosphorus in the phosphodiester bond of the sugar-phosphate backbone of the RNA. There is a transition state (shown above), where the phosphorus is bonded to five oxygen atoms. The phosphorus then detaches from the oxygen connecting it to the adjacent sugar, resulting in ester cleavage of the RNA backbone. (This mechanism is also referred to as RNA cleavage.) This produces a 2’,3’-cyclic phosphate that can then yield either ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or Directed evolution, ''in vitro'' evolved ribozymes are the cleavage (or Ligation (molecular biology), ligation) of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligation (molecular Biology)
Ligation is the joining of two nucleotides, or two nucleic acid fragments, into a single polymeric chain through the action of an enzyme known as a ligase. The reaction involves the formation of a phosphodiester bond between the 3'-hydroxyl terminus of one nucleotide and the 5'-phosphoryl terminus of another nucleotide, which results in the two nucleotides being linked consecutively on a single strand. Ligation works in fundamentally the same way for both DNA and RNA. A cofactor is generally involved in the reaction, usually Adenosine triphosphate, ATP or Nicotinamide riboside, NAD+. Eukaryotic ligases belong to the ATP type, while the NAD+ type are found in bacteria (e.g. Escherichia coli, ''E. coli''). Ligation occurs naturally as part of numerous cellular processes, including DNA replication, transcription, splicing, and recombination, and is also an essential laboratory procedure in molecular cloning, whereby DNA fragments are joined to create recombinant DNA molecules (such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bond Cleavage
In chemistry, bond cleavage, or bond fission, is the splitting of chemical bonds. This can be generally referred to as dissociation when a molecule is cleaved into two or more fragments. In general, there are two classifications for bond cleavage: ''homo''lytic and ''hetero''lytic, depending on the nature of the process. The triplet and singlet excitation energies of a sigma bond can be used to determine if a bond will follow the homolytic or heterolytic pathway. A metal−metal sigma bond is an exception because the bond's excitation energy is extremely high, thus cannot be used for observation purposes. In some cases, bond cleavage requires catalysts. Due to the high bond-dissociation energy of C−H bonds, around , a large amount of energy is required to cleave the hydrogen atom from the carbon and bond a different atom to the carbon. Homolytic cleavage In homolytic cleavage, or homolysis, the two electrons in a cleaved covalent bond are divided equally betwee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Philipp Holliger
Philipp Holliger is a Swiss molecular biologist best known for his work on xeno nucleic acids (XNAs) and RNA engineering. Holliger is a program leader at the MRC Laboratory of Molecular Biology (MRC LMB). Background He earned his degree in Natural Sciences (Dipl. Natwiss. ETH) from ETH Zürich, Switzerland, where he worked with Steven Benner, and his Ph.D. in Molecular Biology at the MRC Centre for Protein Engineering (CPE) in Cambridge under the mentorship of Sir Gregory Winter (CPE and MRC LMB) and Tim Richmond (ETH). While in the Winter laboratory, Holliger developed a new type of bispecific antibody fragment, called a diabody and worked on elucidating the infection pathway of filamentous bacteriophages. After he became an independent group leader at the MRC LMB, Holliger shifted his research focus towards synthetic biology, where he developed methods for emulsion-PCR and ''in vitro'' evolution. Holliger was elected a member of EMBO in 2015. Research XNAs Combining ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |