|
Myc
''Myc'' is a family of regulator genes and proto-oncogenes that code for transcription factors. The ''Myc'' family consists of three related human genes: ''c-myc'' (MYC), ''l-myc'' ( MYCL), and ''n-myc'' (MYCN). ''c-myc'' (also sometimes referred to as ''MYC'') was the first gene to be discovered in this family, due to homology with the viral gene ''v-myc''. In cancer, ''c-myc'' is often constitutively (persistently) expressed. This leads to the increased expression of many genes, some of which are involved in cell proliferation, contributing to the formation of cancer. A common human translocation involving ''c-myc'' is critical to the development of most cases of Burkitt lymphoma. Constitutive upregulation of ''Myc'' genes have also been observed in carcinoma of the cervix, colon, breast, lung and stomach. Myc is thus viewed as a promising target for anti-cancer drugs. Unfortunately, Myc possesses several features that render it undruggable such that any anti-cancer dru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAX (gene)
''MAX'' (also known as myc-associated factor X) is a gene that in humans encodes the MAX transcription factor. Function The protein product of ''MAX'' contains the basic helix-loop-helix and leucine zipper motifs. It is therefore included in the bHLHZ family of transcription factors. It is able to form homodimers with other MAX proteins and heterodimers with other transcription factors, including Mad, Mxl1 and Myc. The homodimers and heterodimers compete for a common DNA target site (the E-box) in a gene promoter zone. Rearrangement of dimers (e.g., Mad:Max, Max:Myc) provides a system of transcriptional regulation with greater diversity of gene targets. Max must dimerise in order to be biologically active. Transcriptionally active hetero- and homodimers involving Max can promote cell proliferation as well as apoptosis. Interactions The protein product of Max has been shown to interact with: * Myc, * MNT, * MSH2, * MXD1, * MXI1, * MYCL1, * N-Myc, * SPAG9, * TEAD1, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-Myc
N-myc proto-oncogene protein also known as N-Myc or basic helix-loop-helix protein 37 (bHLHe37), is a protein that in humans is encoded by the ''MYCN'' gene. Function The ''MYCN'' gene is a member of the MYC family of transcription factors and encodes a protein with a basic helix-loop-helix ( bHLH) domain. This protein is located in the cell nucleus and must dimerize with another bHLH protein in order to bind DNA. N-Myc is highly expressed in the fetal brain and is critical for normal brain development. The ''MYCN'' gene has an antisense RNA, N-cym or ''MYCNOS'', transcribed from the opposite strand which can be translated to form a protein product. N-Myc and ''MYCNOS'' are co-regulated both in normal development and in tumor cells, so it is possible that the two transcripts are functionally related. It has been shown that the antisense RNA encodes for a protein, named NCYM, that has originated ''de novo'' and is specific to human and chimpanzee. This NCYM protein inhibits GSK ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E-box
An E-box (enhancer box) is a DNA response element found in some eukaryotes that acts as a protein-binding site and has been found to regulate gene expression in neurons, muscles, and other tissues. Its specific DNA sequence, CANNTG (where N can be any nucleotide), with a palindromic canonical sequence of CACGTG, is recognized and bound by transcription factors to initiate gene transcription. Once the transcription factors bind to the promoters through the E-box, other enzymes can bind to the promoter and facilitate transcription from DNA to mRNA. Discovery The E-box was discovered in a collaboration between Susumu Tonegawa's and Walter Gilbert's laboratories in 1985 as a control element in immunoglobulin heavy-chain enhancer. They found that a region of 140 base pairs in the tissue-specific transcriptional enhancer element was sufficient for different levels of transcription enhancement in different tissues and sequences. They suggested that proteins made by specific tissue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MYCL
L-myc-1 proto-oncogene protein is a protein that in humans is encoded by the ''MYCL1'' gene. MYCL1 is a bHLH (basic helix-loop-helix) transcription factor implicated in lung cancer. Interactions MYCL1 has been shown to interact Advocates for Informed Choice, doing business as, dba interACT or interACT Advocates for Intersex Youth, is a 501(c)(3) nonprofit organization using innovative strategies to advocate for the legal and human rights of children with intersex trai ... with MAX. References Further reading * * * * * * * * * * * * * * * External links * Transcription factors {{gene-1-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization ( body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Burkitt Lymphoma
Burkitt lymphoma is a cancer of the lymphatic system, particularly B lymphocytes found in the germinal center. It is named after Denis Parsons Burkitt, the Irish surgeon who first described the disease in 1958 while working in equatorial Africa. The overall cure rate for Burkitt lymphoma in developed countries is about 90%, and it is worse in low-income countries. Burkitt lymphoma is uncommon in adults, in whom it has a worse prognosis. Classification Burkitt lymphoma can be divided into three main clinical variants: the endemic, the sporadic, and the immunodeficiency-associated variants. By morphology (i.e., microscopic appearance), immunophenotype, and genetics, the variants of Burkitt lymphoma are alike. * The endemic variant (also called "African variant") most commonly occurs in children living in malaria-endemic regions of the world (e.g., equatorial Africa, Brazil, and Papua New Guinea). Epstein–Barr virus (EBV) infection is found in nearly all patients. Chronic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Burkitt's Lymphoma
Burkitt lymphoma is a cancer of the lymphatic system, particularly B lymphocytes found in the germinal center. It is named after Denis Parsons Burkitt, the Irish surgeon who first described the disease in 1958 while working in equatorial Africa. The overall cure rate for Burkitt lymphoma in developed countries is about 90%, and it is worse in low-income countries. Burkitt lymphoma is uncommon in adults, in whom it has a worse prognosis. Classification Burkitt lymphoma can be divided into three main clinical variants: the endemic, the sporadic, and the immunodeficiency-associated variants. By morphology (i.e., microscopic appearance), immunophenotype, and genetics, the variants of Burkitt lymphoma are alike. * The endemic variant (also called "African variant") most commonly occurs in children living in malaria-endemic regions of the world (e.g., equatorial Africa, Brazil, and Papua New Guinea). Epstein–Barr virus (EBV) infection is found in nearly all patients. Chronic mal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
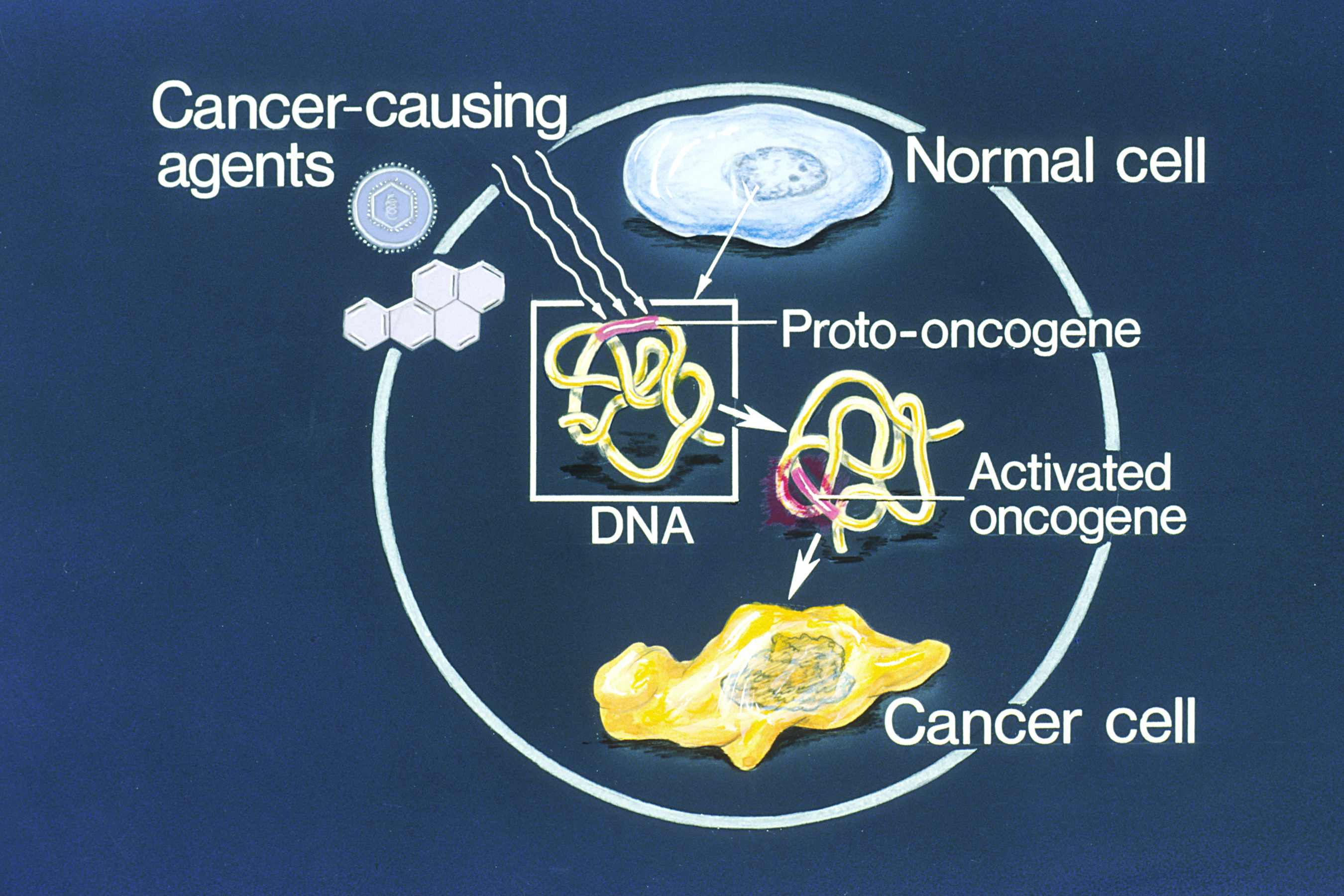
Proto-oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells will undergo a programmed form of rapid cell death ( apoptosis) when critical functions are altered and malfunctioning. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they will predispose the cell to cancer; thus, th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Code
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Effor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine Zipper
A leucine zipper (or leucine scissors) is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization. Leucine zippers are a dimerization motif of the bZIP (Basic-region leucine zipper) class of eukaryotic transcription factors. The bZIP domain is 60 to 80 amino acids in length with a highly conserved DNA binding basic region and a more diversified leuci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme ( RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
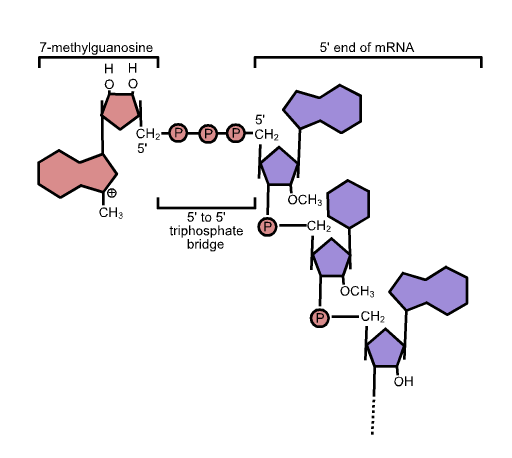
5' Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2′-hydroxy grou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |