|
Myc
''Myc'' is a family of regulator genes and proto-oncogenes that code for transcription factors. The ''Myc'' family consists of three related human genes: ''c-myc'' ( MYC), ''l-myc'' ( MYCL), and ''n-myc'' ( MYCN). ''c-myc'' (also sometimes referred to as ''MYC'') was the first gene to be discovered in this family, due to homology with the viral gene ''v-myc''. In cancer, ''c-myc'' is often constitutively (persistently) expressed. This leads to the increased expression of many genes, some of which are involved in cell proliferation, contributing to the formation of cancer. A common human translocation involving ''c-myc'' is critical to the development of most cases of Burkitt lymphoma. Constitutive upregulation of ''Myc'' genes have also been observed in carcinoma of the cervix, colon, breast, lung and stomach. Myc is thus viewed as a promising target for anti-cancer drugs. Unfortunately, Myc possesses several features that have rendered it difficult to drug to date, such ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAX (gene)
''MAX'' (also known as myc-associated factor X) is a gene that in humans encodes the MAX transcription factor. Function The protein product of ''MAX'' contains the basic helix-loop-helix and leucine zipper motifs. It is therefore included in the bHLHZ family of transcription factors. It is able to form homodimers with other MAX proteins and heterodimers with other transcription factors, including Mad, Mxl1 and Myc. The homodimers and heterodimers compete for a common DNA target site (the E-box) in a gene promoter zone. Rearrangement of dimers (e.g., Mad:Max, Max:Myc) provides a system of transcriptional regulation with greater diversity of gene targets. Max must dimerise in order to be biologically active. Transcriptionally active hetero- and homodimers involving Max can promote cell proliferation as well as apoptosis. Interactions The protein product of Max has been shown to interact with: * Myc, * MNT, * MSH2, * MXD1, * MXI1, * MYCL1, * N-Myc, * SPAG9, * T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Reprogramming
Induced pluripotent stem cells (also known as iPS cells or iPSCs) are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Japan, who together showed in 2006 that the introduction of four specific genes (named Myc, Oct3/4, Sox2 and Klf4), collectively known as Yamanaka factors, encoding transcription factors could convert somatic cells into pluripotent stem cells. Shinya Yamanaka was awarded the 2012 Nobel Prize along with Sir John Gurdon "for the discovery that mature cells can be reprogrammed to become pluripotent." Pluripotent stem cells hold promise in the field of regenerative medicine. Because they can propagate indefinitely, as well as give rise to every other cell type in the body (such as neurons, heart, pancreatic, and liver cells), they represent a single source of cells that could be used to replace those lost to damage or disease. The most wel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Induced Pluripotent Stem Cells
Induced pluripotent stem cells (also known as iPS cells or iPSCs) are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Japan, who together showed in 2006 that the introduction of four specific genes (named Myc, Oct3/4, Sox2 and Klf4), collectively known as Yamanaka factors, encoding transcription factors could convert somatic cells into pluripotent stem cells. Shinya Yamanaka was awarded the 2012 Nobel Prize along with Sir John Gurdon "for the discovery that mature cells can be reprogrammed to become pluripotent." Pluripotent stem cells hold promise in the field of regenerative medicine. Because they can propagate indefinitely, as well as give rise to every other cell type in the body (such as neurons, heart, pancreatic, and liver cells), they represent a single source of cells that could be used to replace those lost to damage or disease. The most w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-Myc
N-myc proto-oncogene protein also known as N-Myc or basic helix-loop-helix protein 37 (bHLHe37), is a protein that in humans is encoded by the ''MYCN'' gene. Function The ''MYCN'' gene is a member of the MYC family of transcription factors and encodes a protein with a basic helix-loop-helix ( bHLH) domain. This protein is located in the cell nucleus and must dimerize with another bHLH protein in order to bind DNA. N-Myc is highly expressed in the fetal brain and is critical for normal brain development. The ''MYCN'' gene has an antisense RNA, N-cym or ''MYCNOS'', transcribed from the opposite strand which can be translated to form a protein product. N-Myc and ''MYCNOS'' are co-regulated both in normal development and in tumor cells, so it is possible that the two transcripts are functionally related. It has been shown that the antisense RNA encodes for a protein, named NCYM, that has originated ''de novo'' and is specific to human and chimpanzee. This NCYM protein inhibits GSK ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E-box
An E-box (enhancer box) is a Response element, DNA response element found in some eukaryotes that acts as a protein-binding site and has been found to regulate gene expression in neurons, muscles, and other tissues. Its specific DNA sequence, CANNTG (where N can be any nucleotide), with a palindromic canonical sequence of CACGTG, is recognized and bound by transcription factors to initiate gene transcription (genetics), transcription. Once the transcription factors bind to the promoters through the E-box, other enzymes can bind to the promoter and facilitate transcription from DNA to mRNA. Discovery The E-box was discovered in a collaboration between Susumu Tonegawa's and Walter Gilbert's laboratories in 1985 as a control element in immunoglobulin heavy-chain enhancer. They found that a region of 140 base pairs in the tissue-specific transcriptional enhancer element was sufficient for different levels of transcription Enhancer (genetics), enhancement in different tissues and sequenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MYCL
L-myc-1 proto-oncogene protein is a protein that in humans is encoded by the ''MYCL1'' gene. MYCL1 is a bHLH (basic helix-loop-helix) transcription factor implicated in lung cancer. Interactions MYCL1 has been shown to interact with MAX Max or MAX may refer to: Animals * Max (American dog) (1983–2013), at one time purported to be the world's oldest living dog * Max (British dog), the first pet dog to win the PDSA Order of Merit (animal equivalent of the OBE) * Max (gorilla) .... References Further reading * * * * * * * * * * * * * * * External links * Transcription factors {{gene-1-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are Gene expression, expressed in the desired Cell (biology), cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
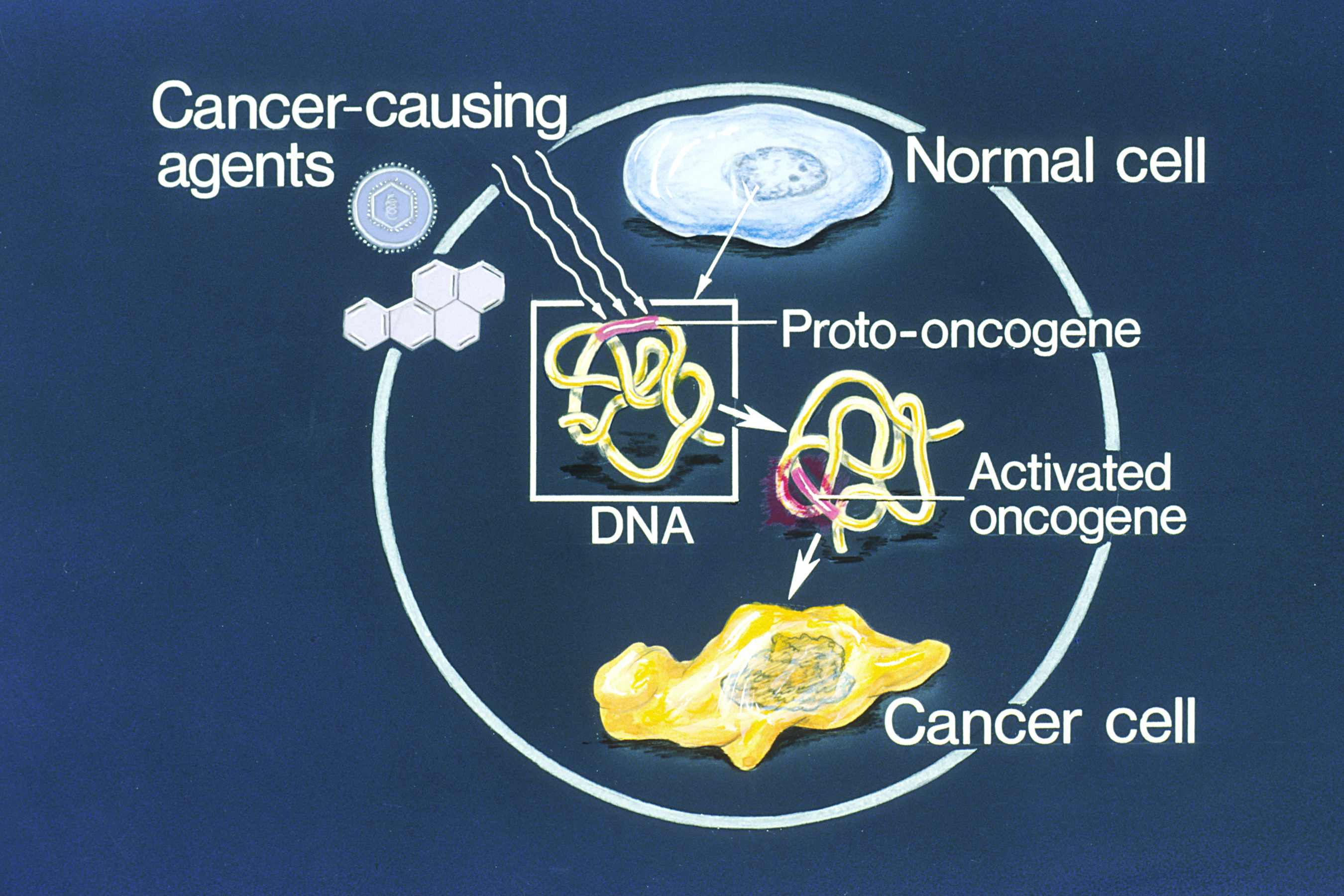
Proto-oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells undergo a preprogrammed rapid cell death () if critical functions are altered and then malfunction. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they predispose the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yamanaka Factors
is a Japanese stem cell researcher and a Nobel Prize laureate. He is a professor and the director emeritus of Center for iPS Cell ( induced Pluripotent Stem Cell) Research and Application, Kyoto University; as a senior investigator at the UCSF-affiliated Gladstone Institutes in San Francisco, California; and as a professor of anatomy at University of California, San Francisco (UCSF). Yamanaka is also a past president of the International Society for Stem Cell Research (ISSCR). He received the 2010 BBVA Foundation Frontiers of Knowledge Award in the biomedicine category, the 2011 Wolf Prize in Medicine with Rudolf Jaenisch, and the 2012 Millennium Technology Prize together with Linus Torvalds. In 2012, he and John Gurdon were awarded the Nobel Prize for Physiology or Medicine for the discovery that mature cells can be converted to stem cells. In 2013, he was awarded the $3 million Breakthrough Prize in Life Sciences for his work. Education Yamanaka was born in H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem Cell
In multicellular organisms, stem cells are undifferentiated or partially differentiated cells that can change into various types of cells and proliferate indefinitely to produce more of the same stem cell. They are the earliest type of cell in a cell lineage. They are found in both embryonic and adult organisms, but they have slightly different properties in each. They are usually distinguished from progenitor cells, which cannot divide indefinitely, and precursor or blast cells, which are usually committed to differentiating into one cell type. In mammals, roughly 50 to 150 cells make up the inner cell mass during the blastocyst stage of embryonic development, around days 5–14. These have stem-cell capability. '' In vivo'', they eventually differentiate into all of the body's cell types (making them pluripotent). This process starts with the differentiation into the three germ layers – the ectoderm, mesoderm and endoderm – at the gastrulation stage. However, whe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the Codon tables, RNA codon table). That scheme is often called the canonical or standard genetic code, or simply ''the'' genetic code, though #Variations, variant codes (suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Discovery
Discovery may refer to: * Discovery (observation), observing or finding something unknown * Discovery (fiction), a character's learning something unknown * Discovery (law), a process in courts of law relating to evidence Discovery, The Discovery or Discoveries may also refer to: Film and television * The Discovery (film), ''The Discovery'' (film), a 2017 British-American romantic science fiction film * Discovery Channel, an American TV channel owned by Warner Bros. Discovery * Discovery (Canadian TV series), ''Discovery'' (Canadian TV series), a 1962–1963 Canadian documentary television program * Discovery (Irish TV series), ''Discovery'' (Irish TV series), an Irish documentary television programme * Discovery (UK TV programme), ''Discovery'' (UK TV programme), a British documentary television programme * Discovery (U.S. TV series), ''Discovery'' (U.S. TV series), a 1962–1971 American television news program * ''Star Trek: Discovery'', an American television series ** USS D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |