|
Human Accelerated Regions
Human accelerated regions (HARs), first described in August 2006, are a set of 49 segments of the human genome that are conserved throughout vertebrate evolution but are strikingly different in humans. They are named according to their degree of difference between humans and chimpanzees (HAR1 showing the largest degree of human-chimpanzee differences). Found by scanning through genomic databases of multiple species, some of these highly mutated areas may contribute to human-specific traits. Others may represent loss of functional mutations, possibly due to the action of biased gene conversion rather than adaptive evolution. Several of the HARs encompass genes known to produce proteins important in neurodevelopment. HAR1 is a 106-base pair stretch found on the long arm of chromosome 20 overlapping with part of the RNA genes HAR1F and HAR1R. HAR1F is active in the developing human brain. The HAR1 sequence is found (and conserved) in chickens and chimpanzees but is not present in fish ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences. Introns make up a large percentage of non-coding DNA. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thumb
The thumb is the first digit of the hand, next to the index finger. When a person is standing in the medical anatomical position (where the palm is facing to the front), the thumb is the outermost digit. The Medical Latin English noun for thumb is ''pollex'' (compare ''hallux'' for big toe), and the corresponding adjective for thumb is ''pollical''. Definition Thumb and fingers The English word ''finger'' has two senses, even in the context of appendages of a single typical human hand: # Any of the five terminal members of the hand. # Any of the four terminal members of the hand, other than the thumb Linguistically, it appears that the original sense was the first of these two: (also rendered as ) was, in the inferred Proto-Indo-European language, a suffixed form of (or ), which has given rise to many Indo-European-family words (tens of them defined in English dictionaries) that involve, or stem from, concepts of fiveness. The thumb shares the following with each of the o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FHIT
Bis(5'-adenosyl)-triphosphatase also known as fragile histidine triad protein (FHIT) is an enzyme that in humans is encoded by the ''FHIT'' gene. Function FHIT is also known as human accelerated region 10. It may, therefore, have played a key role in differentiating humans from apes. This gene, a member of the histidine triad gene family, encodes a diadenosine P1,P3-bis(5'-adenosyl)-triphosphate adenylohydrolase involved in purine metabolism. The gene encompasses the common fragile site FRA3B on chromosome 3, where carcinogen-induced damage can lead to translocations and aberrant transcripts of this gene. In fact, aberrant transcripts from this gene have been found in about half of all esophageal, stomach, and colon carcinomas. Though the exact molecular function of FHIT is still partially unclear, the gene works as a tumor suppressor as it has been demonstrated in animal studies. Furthermore FHIT has been shown to synergize with VHL, another tumor suppressor, in protectin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PTPRT
Receptor-type tyrosine-protein phosphatase T is an enzyme that in humans is encoded by the ''PTPRT'' gene. PTPRT is also known as PTPrho, PTPρ and human accelerated region 9. The human accelerated regions are 49 regions of the human genome that are conserved among vertebrates, but in humans show significant distinction from other vertebrates. This region may, therefore, have played a key role in differentiating humans from apes. Function The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. PTPrho has been proposed to function during development of the nervous system and as a tumor suppressor in cancer. Structure This PTP possesses an extracellular region, a single transmembrane region, and two tandem intracellular catalytic domains, and thus represents a recept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
POU Family
POU or pou may refer to: People * Pou (surname), a surname * Chu Pou (303–350), Chinese general and politician * Pou Temara (born 1948), New Zealand Māori academic Codes * POU, IATA airport code and FAA location identifier for Hudson Valley Regional Airport, New York, United States * POU, Amtrak station code for Poughkeepsie station, a rail station in Poughkeepsie, New York, United States * pou, deprecated ISO 639-3 code for the Southern Poqomam language, spoken in Guatemala Other uses * ''Pou'' (video game), a 2012 video game * Pou (deity), a Moriori deity. * POU domain POU (pronounced 'pow') is a family of proteins that have well-conserved homeodomains. Etymology The acronym POU is derived from the names of three transcription factors: * the Pituitary-specific Pit-1 * the Octamer transcription factor prot ..., the conserved region in the POU family of proteins * Point of use (POU) water treatment equipment, also called portable water purification {{disambigua ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WWOX
WW domain-containing oxidoreductase is an enzyme that in humans is encoded by the ''WWOX'' gene. Function WW domain-containing proteins are found in all eukaryotes and play an important role in the regulation of a wide variety of cellular functions such as protein degradation, transcription, and RNA splicing. This gene encodes a protein which contains 2 WW domains and a short-chain dehydrogenase/reductase domain (SRD). The highest normal expression of this gene is detected in hormonally regulated tissues such as testis, ovary, and prostate. This expression pattern and the presence of an SRD domain suggest a role for this gene in steroid metabolism. The encoded protein is more than 90% identical to the mouse protein, which is an essential mediator of tumor necrosis factor-alpha-induced apoptosis, suggesting a similar, important role in apoptosis for the human protein. In addition, there is evidence that this gene behaves as a suppressor of tumor growth. Alternative splicing of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
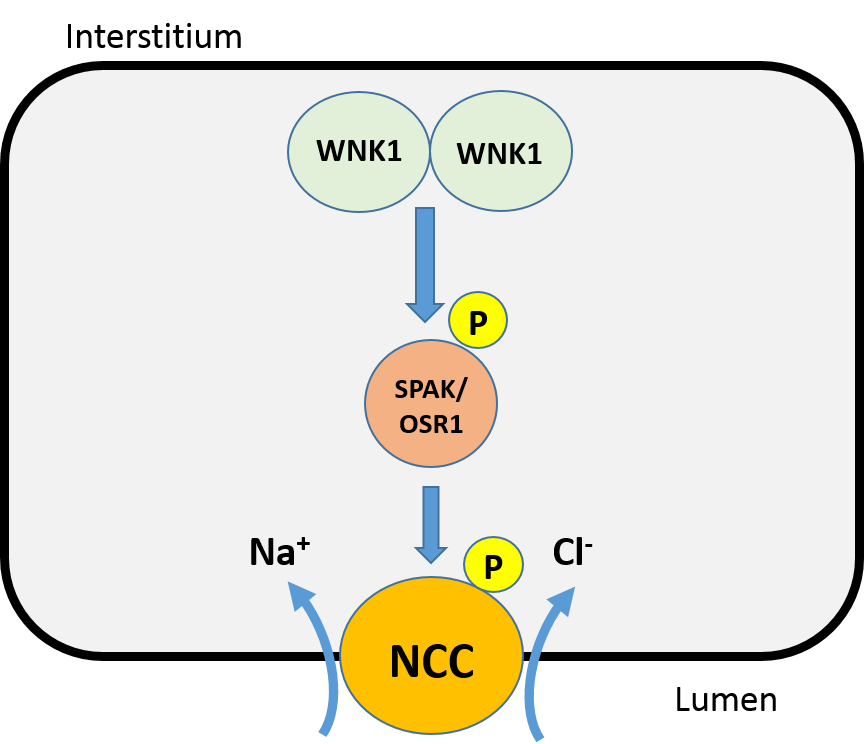
WNK1
WNK (lysine deficient protein kinase 1), also known as WNK1, is an enzyme that is encoded by the ''WNK1'' gene. WNK1 is serine-threonine protein kinase and part of the "with no lysine/K" kinase WNK family. The predominant role of WNK1 is the regulation of cation-Cl− cotransporters (CCCs) such as the sodium chloride cotransporter ( NCC), basolateral Na-K-Cl symporter (NKCC1), and potassium chloride cotransporter (KCC1) located within the kidney. CCCs mediate ion homeostasis and modulate blood pressure by transporting ions in and out of the cell. ''WNK1'' mutations as a result have been implicated in blood pressure disorders/diseases; a prime example being familial hyperkalemic hypertension (FHHt). Structure The WNK1 protein is composed of 2382 amino acids (molecular weight 230 kDa). The protein contains a kinase domain located within its short N-terminal domain and a long C-terminal tail. The kinase domain has some similarity to the MEKK protein kinase family. As a member ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAD1L1
Mitotic spindle assembly checkpoint protein MAD1 is a protein that in humans is encoded by the ''MAD1L1'' gene. MAD1L1 is also known as Human Accelerated Region 3. It may have played a key role in the evolution of humans from apes. Function MAD1L1 is a component of the mitotic spindle-assembly checkpoint that prevents the onset of anaphase until all chromosome are properly aligned at the metaphase plate. MAD1L1 functions as a homodimer and interacts with MAD2L1. MAD1L1 may play a role in cell cycle control and tumor suppression. Three transcript variants encoding the same protein have been found for this gene. Interactions MAD1L1 has been shown to interact with: * HDAC1, * Histone deacetylase 2, and * MAD2L1, See also * MAD1 * MAD2 * Hyperphosphorylation Hyperphosphorylation occurs when a biochemical with multiple phosphorylation sites is fully saturated. Hyperphosphorylation is one of the signaling mechanisms used by the cell to regulate mitosis. When these mechanis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CENTG2
Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 1 is an enzyme that in humans is encoded by the ''AGAP1'' gene. Function CENTG2 belongs to an ADP-ribosylation factor GTPase-activating (ARF-GAP) protein family involved in membrane traffic and actin cytoskeleton dynamics (Nie et al., 2002). upplied by OMIMref name="entrez" /> HACNS1 HACNS1 is located in an intron of the gene CENTG2 (also known as Human Accelerated Region 2). HACNS1 is hypothesized to be a gene enhancer "that may have contributed to the evolution of the uniquely opposable human thumb, and possibly also modifications in the ankle or foot that allow humans to walk on two legs". Evidence to date shows that of the 110,000 gene enhancer sequences identified in the human genome, HACNS1 has undergone the most change during the evolution of humans following the split with the ancestors of chimpanzees. Model organisms Model organisms have been used in the study of AGAP1 function. A conditional knockout ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HAR1F & HAR1R
In molecular biology, Human accelerated region 1 (highly accelerated region 1, HAR1) is a segment of the human genome found on the long arm of chromosome 20. It is a Human accelerated region. It is located within a pair of overlapping long non-coding RNA genes, HAR1A (HAR1F) and HAR1B (HAR1R). HAR1A HAR1A is expressed in Cajal-Retzius cells, contemporaneously with the protein reelin. HAR1A was identified in August 2006 when human accelerated regions (HARs) were first investigated. These 49 regions represent parts of the human genome that differ significantly from highly conserved regions of our closest ancestors in terms of evolution. Many of the HARs are associated with genes known to play a role in neurodevelopment. One particularly altered region, HAR1, was found in a stretch of genome with no known protein-coding RNA sequences. Two RNA genes, HAR1F and HAR1R, were identified partly within the region. The RNA structure of HAR1A has been shown to be stable, with a secondary str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation tends to exist within any given population as a result of genetic mutation and recombination. Evolution occurs when evolutionary processes such as natural selection (including sexual selection) and genetic drift act on this variation, resulting in certain characteristics becoming more common or more rare within a population. The evolutionary pressures that determine whether a characteristic is common or rare within a population constantly change, resulting in a change in heritable characteristics arising over successive generations. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms, and molecules. The theory of evolution by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |