|
Gap Gene
A gap gene is a type of gene involved in the development of the segmented embryos of some arthropods. Gap genes are defined by the effect of a mutation in that gene, which causes the loss of contiguous body segments, resembling a gap in the normal body plan. Each gap gene, therefore, is necessary for the development of a section of the organism. Gap genes were first described by Christiane Nüsslein-Volhard and Eric Wieschaus in 1980. They used a genetic screen to identify genes required for embryonic development in the fruit fly ''Drosophila melanogaster''. They found three genes – ''knirps, Krüppel and hunchback'' – where mutations caused deletion of particular stretches of segments. Later work identified more gap genes in the ''Drosophila'' early embryo – ''giant'', ''huckebein'' and ''tailless''. Further gap genes including orthodenticle and buttonhead are required for the development of the ''Drosophila'' head. Once the gap genes had been identified at the molecu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Krüppel
Krüppel is a gap gene in ''Drosophila melanogaster'', located on the 2R chromosome, which encodes a zinc finger C2H2 transcription factor. Gap genes work together to establish the anterior-posterior segment patterning of the insect through regulation of the transcription factor encoding pair rule genes. These genes in turn regulate segment polarity genes. ''Krüppel'' means "cripple" in German, named for the crippled appearance of mutant larvae, who have failed to develop proper thoracic and anterior segments in the abdominal region. Mutants can also have abdominal mirror duplications. Human homologs of Krüppel are collectively named ''Krüppel''-like factors, a set of proteins well characterized for their role in carcinogenesis. ''Krüppel'' expression pathway ''Krüppel'' is expressed in the center of the embryo during the cellular blastoderm stage of development. Its expression pattern is restricted to this domain largely through interactions with the maternal effect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pair-rule Gene
A pair-rule gene is a type of gene involved in the development of the segmented embryos of insects. Pair-rule genes are expressed as a result of differing concentrations of gap gene proteins, which encode transcription factors controlling pair-rule gene expression. Pair-rule genes are defined by the effect of a mutation in that gene, which causes the loss of the normal developmental pattern in alternating segments. Pair-rule genes were first described by Christiane Nüsslein-Volhard and Eric Wieschaus in 1980. They used a genetic screen to identify genes required for embryonic development in the fruit fly ''Drosophila melanogaster''. In normal unmutated ''Drosophila,'' each segment produces bristles called denticles in a band arranged on the side of the segment closer to the head (the anterior Standard anatomical terms of location are used to describe unambiguously the anatomy of humans and other animals. The terms, typically derived from Latin or Greek roots, describe som ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
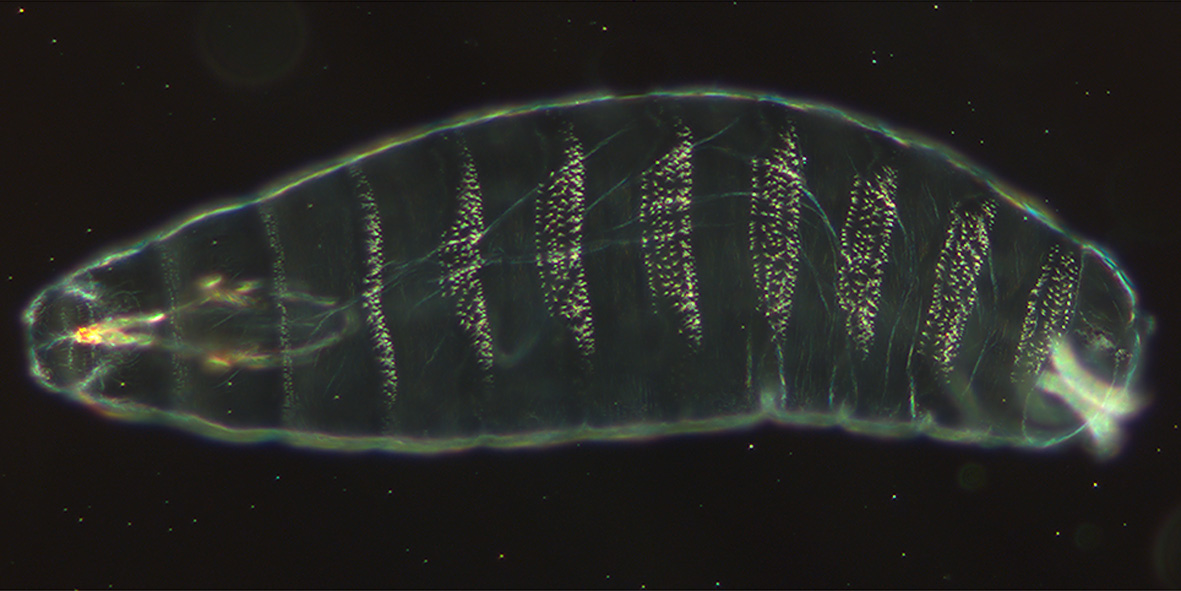
Drosophila Embryogenesis
''Drosophila'' embryogenesis, the process by which ''Drosophila'' (fruit fly) embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. ''Drosophila melanogaster'' was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909. Life cycle ''Drosophila'' display a holometabolous method of development, meaning that they have three distinct stages of their post-embryonic life cycle, each with a radically different body plan: larva, pupa and finally, adult. The machinery necessary for the function and smooth transition between these three phases develops during embryogenesis. During embryogenesis, the larval stage fly will develop and ha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bicoid
Homeotic protein bicoid is encoded by the ''bcd'' maternal effect gene in ''Drosophilia''. Homeotic protein bicoid concentration gradient patterns the anterior-posterior (A-P) axis during ''Drosophila'' embryogenesis. Bicoid was the first protein demonstrated to act as a morphogen. Although bicoid is important for the development of ''Drosophila'' and other higher dipterans, it is absent from most other insects, where its role is accomplished by other genes. Role in axial patterning ''Bicoid'' mRNA is actively localized to the anterior of the fruit fly egg during oogenesis along microtubules by the motor protein dynein, and retained there through association with cortical actin. Translation of ''bicoid'' is regulated by its 3′ UTR and begins after egg deposition. Diffusion and convection within the syncytium produce an exponential gradient of Bicoid protein within roughly one hour, after which Bicoid nuclear concentrations remain approximately constant through cellulariza ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maternal Effect
A maternal effect is a situation where the phenotype of an organism is determined not only by the environment it experiences and its genotype, but also by the environment and genotype of its mother. In genetics, maternal effects occur when an organism shows the phenotype expected from the genotype of the mother, irrespective of its own genotype, often due to the mother supplying messenger RNA or proteins to the egg. Maternal effects can also be caused by the maternal environment independent of genotype, sometimes controlling the size, sex, or behaviour of the offspring. These adaptive maternal effects lead to phenotypes of offspring that increase their fitness. Further, it introduces the concept of phenotypic plasticity, an important evolutionary concept. It has been proposed that maternal effects are important for the evolution of adaptive responses to environmental heterogeneity. In genetics In genetics, a maternal effect occurs when the phenotype of an organism is determined by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pair-rule Gene
A pair-rule gene is a type of gene involved in the development of the segmented embryos of insects. Pair-rule genes are expressed as a result of differing concentrations of gap gene proteins, which encode transcription factors controlling pair-rule gene expression. Pair-rule genes are defined by the effect of a mutation in that gene, which causes the loss of the normal developmental pattern in alternating segments. Pair-rule genes were first described by Christiane Nüsslein-Volhard and Eric Wieschaus in 1980. They used a genetic screen to identify genes required for embryonic development in the fruit fly ''Drosophila melanogaster''. In normal unmutated ''Drosophila,'' each segment produces bristles called denticles in a band arranged on the side of the segment closer to the head (the anterior Standard anatomical terms of location are used to describe unambiguously the anatomy of humans and other animals. The terms, typically derived from Latin or Greek roots, describe som ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as Transfer RNA, transfer RNA (tRNA) and Small nuclear RNA, small nuclear RNA (snRNA), the product is a functional List of RNAs, non-coding RNA. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and viruses—to generate the macromolecule, macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, ''i.e.'' observable trait. The genetic information stored in DNA represents the genotype, whereas the phenotype results from the "interpretation" of that informati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthodenticle
Orthodenticle (otd) is a homeobox gene found in Drosophila that regulates the development of anterior patterning, with particular involvement in the central nervous system function and eye development. It is located on the X chromosome. The gene is an ortholog of the human OTX1/ OTX2 gene. Function During embryonic Drosophila development, otd is required for the head and ventral midline to develop correctly. In the larval stage, otd is expressed in specific sac-like epithelial structures known as imaginal discs that later give rise to external structures of the head and thorax. Particularly, otd is required for the development of the dorsal region of the adult ''Drosophila'' head, that forms from the fusion of two eye-antennal discs. Distribution of the otd protein occurs along a concentration of the imaginal disc Primordium, primordia for these head structures such that different levels of otd expression are required for differential mediolateral subdomains to be established. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hunchback (gene)
Hunchback is a maternal effect and zygotic gene expressed in the embryos of the fruit fly ''Drosophila melanogaster''. In maternal effect genes, the RNA or protein from the mother’s gene is deposited into the oocyte or embryo before the embryo can express its own zygotic genes. Hunchback is a morphogen, meaning the concentration gradient of ''Hunchback'' at a specific region determines the segment or body part it develops into. This is possible because ''Hunchback'' is a transcription factor protein that binds to genes’ regulatory regions, changing RNA expression levels. ''Hunchback'' expression pathway Maternal ''Hunchback'' RNA enters the embryo at the syncytial blastoderm stage, where the entire embryo has undergone many nuclear divisions but has one communal cytoplasm, allowing for RNA to disperse freely throughout the embryo. This allows the maternal effect genes ''Hunchback'', ''Bicoid'', ''Nanos'', and ''Caudal'' to regulate zygotic genes to create different identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly", or "banana fly". In the wild, ''D. melanogaster'' are attracted to rotting fruit and fermenting beverages, and are often found in orchards, kitchens and pubs. Starting with Charles W. Woodworth's 1901 proposal of the use of this species as a model organism, ''D. melanogaster'' continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and Life history theory, life history evolution. ''D. melanogaster'' was the first animal to be Fruit flies in space, launched into space in 1947. As of 2017, six Nobel Prizes have been awarded to drosophilists for their work using the insect. ''Drosophila melanogaster'' is typically used in research owing to its rapid life cycle, relatively simple genetics with on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |