|
External Transcribed Spacer
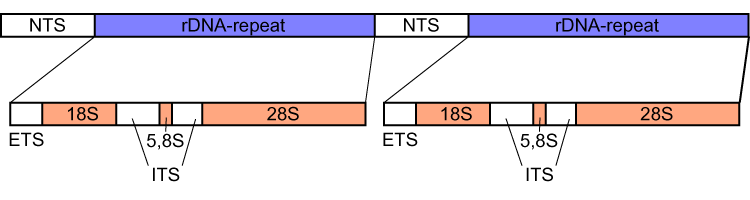
External transcribed spacer (ETS) refers to a piece of non-functional RNA, closely related to the internal transcribed spacer, which is situated outside structural ribosomal RNAs (rRNA) on a common precursor transcript. ETS sequences characteristic to an organism can be used to trace its phylogeny A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or Taxon, taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, M .... References {{genetics-stub DNA Phylogenetics de:Spacer (Biologie) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Internal Transcribed Spacer
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. Across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRNA ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogeny
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or Taxon, taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In other words, it is a branching diagram or a tree (graph theory), tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. In evolutionary biology, all life on Earth is theoretically part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the study of phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. computational phylogenetics, Computational phylogenetics (also phylogeny inference) focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape. Phylogene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical data and observed heritable traits of DNA sequences, protein amino acid sequences, and morphology. The results are a phylogenetic tree—a diagram depicting the hypothetical relationships among the organisms, reflecting their inferred evolutionary history. The tips of a phylogenetic tree represent the observed entities, which can be living taxa or fossils. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the taxa represented on the tree. An unrooted tree diagram (a network) makes no assumption about directionality of character state transformation, and does not show the origin or "root" of the taxa in question. In addition to their use for inferring phylogenetic pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |