|
Exonic Splicing Silencer
An exonic splicing silencer (ESS) is a short region (usually 4-18 nucleotides) of an exon and is a cis-regulatory element. A set of 103 hexanucleotides known as FAS-hex3 has been shown to be abundant in ESS regions. ESSs inhibit or silence splicing of the pre-mRNA and contribute to constitutive and alternate splicing. To elicit the silencing effect, ESSs recruit proteins that will negatively affect the core splicing machinery. Mechanism of action Exonic splicing silencers work by inhibiting the splicing of pre-mRNA strands or promoting exon skipping. The single stranded pre-mRNA molecules need to have their intronic and exonic regions spliced in order to be translated. ESSs silence splice sites adjacent to them by interfering with the components of the core splicing complex, such as the snRNP's, U1 and U2. This causes proteins that negatively influence splicing to be recruited to the splicing machinery. ESSs have four general roles: * inhibiting exon inclusion * inhibiting in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' is a shortening of the phrase ''expressed region'' and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron... must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cystic Fibrosis
Cystic fibrosis (CF) is a genetic disorder inherited in an autosomal recessive manner that impairs the normal clearance of Sputum, mucus from the lungs, which facilitates the colonization and infection of the lungs by bacteria, notably ''Staphylococcus aureus''. CF is a rare genetic disorder that affects mostly the lungs, but also the pancreas, liver, kidneys, and intestine. The hallmark feature of CF is the accumulation of thick mucus in different organs. Long-term issues include Shortness of breath, difficulty breathing and coughing up mucus as a result of frequent pneumonia, lung infections. Other signs and symptoms may include Sinusitis, sinus infections, failure to thrive, poor growth, Steatorrhea, fatty stool, Nail clubbing, clubbing of the fingers and toes, and infertility in most males. Different people may have different degrees of symptoms. Cystic fibrosis is inherited in an autosomal recessive manner. It is caused by the presence of mutations in both copies (alleles) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
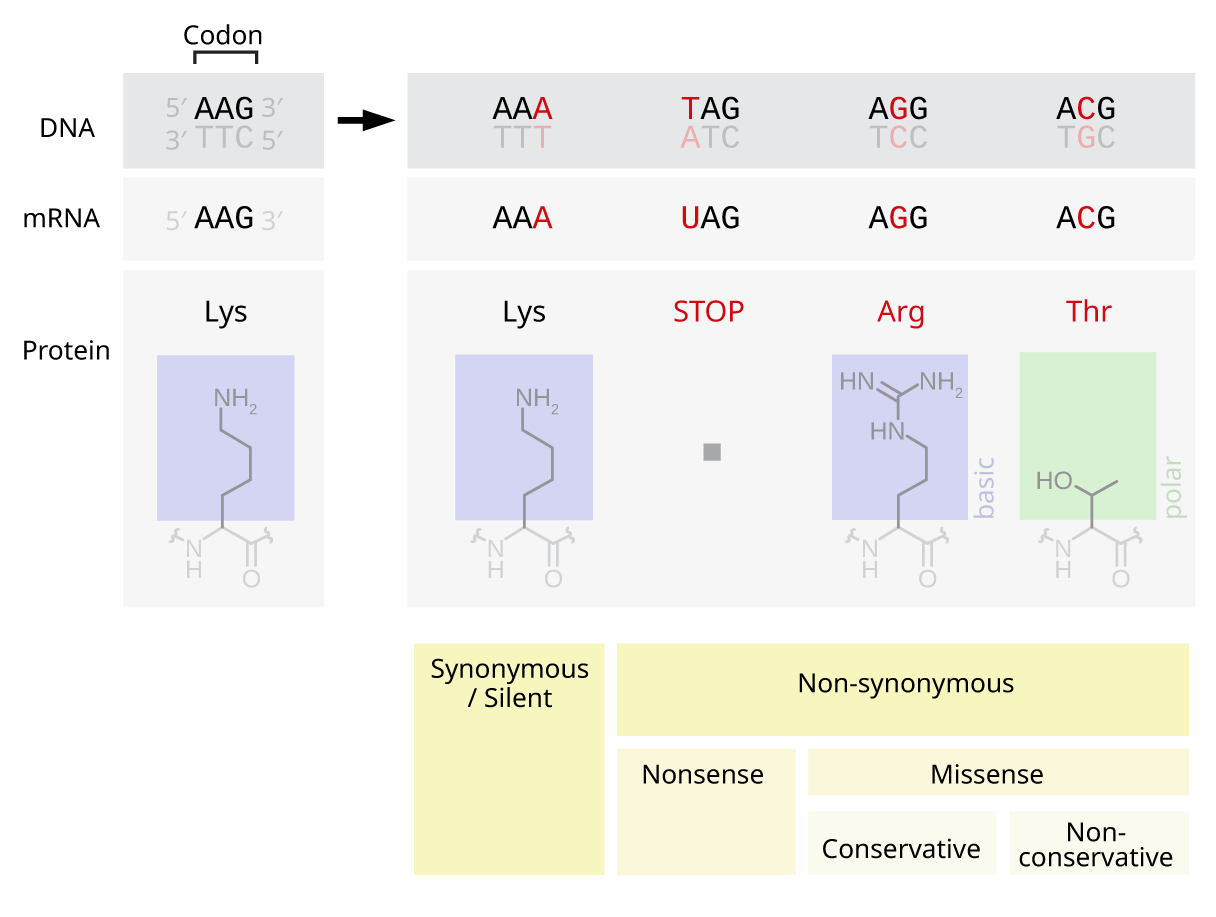
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. Synonymous substitution, synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ataxia Telangiectasia
Ataxia (from Greek α- negative prefix+ -τάξις rder= "lack of order") is a neurological sign consisting of lack of voluntary coordination of muscle movements that can include gait abnormality, speech changes, and abnormalities in eye movements, that indicates dysfunction of parts of the nervous system that coordinate movement, such as the cerebellum. These nervous system dysfunctions occur in several different patterns, with different results and different possible causes. Ataxia can be limited to one side of the body, which is referred to as hemiataxia. Friedreich's ataxia has gait abnormality as the most commonly presented symptom. Dystaxia is a mild degree of ataxia. Types Cerebellar The term cerebellar ataxia is used to indicate ataxia due to dysfunction of the cerebellum. The cerebellum is responsible for integrating a significant amount of neural information that is used to coordinate smoothly ongoing movements and to participate in motor planning. A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
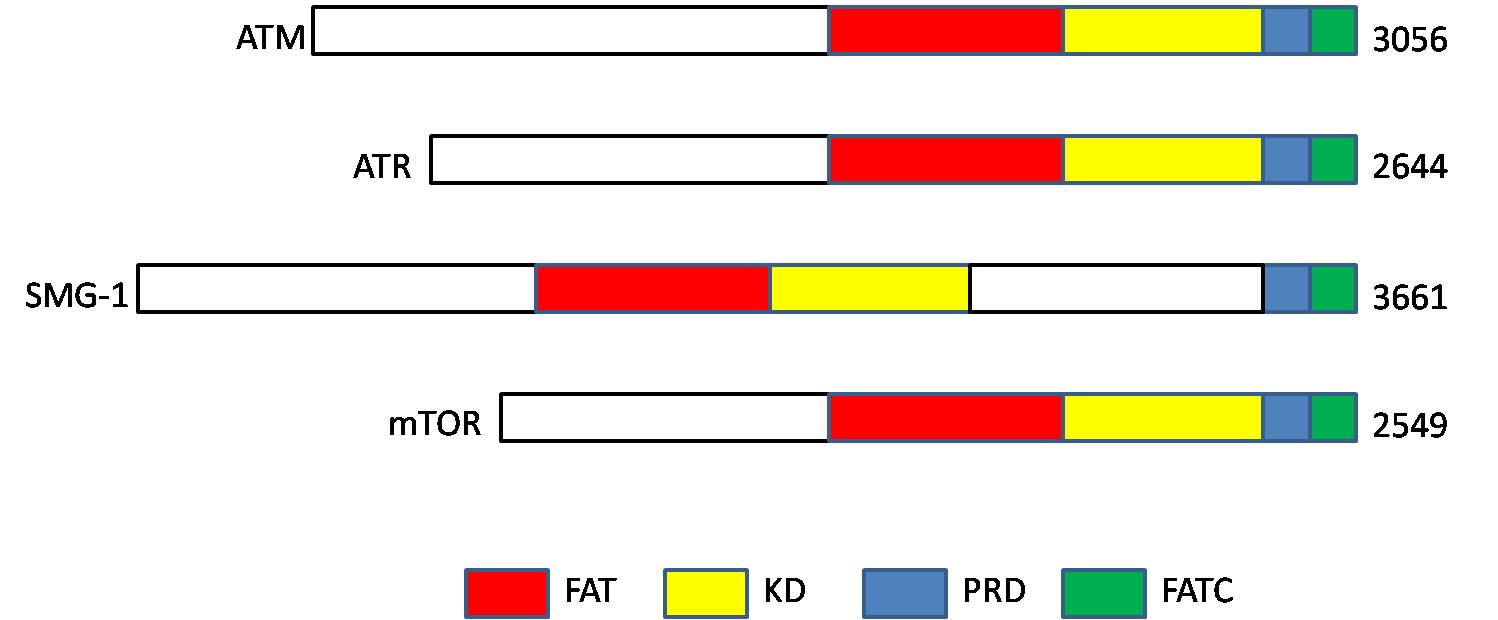
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA repair#Double-strand breaks, DNA double-strand breaks (Canonical pathway, canonical pathway), oxidative stress, topoisomerase cleavage complexes, splicing intermediates, R-loops and in some cases by DNA repair, single-strand DNA breaks. It phosphorylates several key proteins that initiate activation of the DNA damage cell cycle checkpoint, checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exonic Splicing Enhancer
In molecular biology, an exonic splicing enhancer (ESE) is a DNA sequence motif consisting of 6 bases within an exon that directs, or enhances, accurate splicing of heterogeneous nuclear RNA ( hnRNA) or pre-mRNA into messenger RNA (mRNA). Introduction Short sequences of DNA are transcribed to RNA; then this RNA is translated to a protein. A gene located in DNA will contain introns and exons. Part of the process of preparing the RNA includes splicing out the introns, sections of RNA that do not code for the protein. The presence of exonic splicing enhancers is essential for proper identification of splice sites by the cellular machinery. Role in splicing SR proteins bind to and promote exon splicing in regions with ESEs, while heterogeneous ribonucleoprotein particles (hnRNPs) bind to and block exon splicing in regions with exonic splicing silencers. Both types of proteins are involved in the assembly and proper functioning of spliceosomes. During RNA splicing, U2 s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transition (genetics)
Transition, in genetics and molecular biology, refers to a point mutation that changes a purine nucleotide to another purine (Adenine, A ↔ Guanine, G), or a pyrimidine nucleotide to another pyrimidine (Cytosine, C ↔ Thymine, T). Approximately two out of three single nucleotide polymorphisms (SNPs) are transitions. Transitions can be caused by oxidative deamination and tautomerization. Although there are twice as many possible transversions, transitions appear more often in genomes, possibly due to the molecular mechanisms that generate them. Transitions are more likely to be Synonymous substitution, synonymous substitutions than transversions, as one observes in the DNA and RNA codon tables, codon table. 5-Methylcytosine is more prone to transition than unmethylated cytosine, due to spontaneous deamination. This mechanism is important because it dictates the rarity of CpG islands. See also * Transversion References External links Diagram at mun.ca Mutation {{Ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SMN2
Survival of motor neuron 2 (''SMN2'') is a gene that encodes the SMN protein (full and truncated) in humans. Gene The ''SMN2'' gene is part of a 500 kb inverted duplication on chromosome 5q13. This duplicated region contains at least four genes and repetitive elements which make it prone to rearrangements and deletions. The repetitiveness and complexity of the sequence have also caused difficulty in determining the organization of this genomic region. The telomeric (''SMN1'') and centromeric (''SMN2'') copies of this gene are nearly identical and encode the same protein. The critical sequence difference between the two genes is a single nucleotide in exon 7, which is thought to be an exon splice enhancer. The nucleotide substitution in ''SMN2'' results in around 80-90% of its transcripts to be a truncated, unstable protein of no biological function (Δ7SMN) and only 10-20% of its transcripts being full-length protein (fl-SMN). Note that the nine exons of both the telomer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SMN1
Survival of motor neuron 1 (''SMN1''), also known as component of gems 1 or ''GEMIN1'', is a gene that encodes the SMN protein in humans. Gene ''SMN1'' is the telomeric copy of the gene encoding the SMN protein; the centromeric copy is termed '' SMN2''. ''SMN1'' and ''SMN2'' are part of a 500 kbp inverted duplication on chromosome 5q13. This duplicated region contains at least four genes and repetitive elements which make it prone to rearrangements and deletions. The repetitiveness and complexity of the sequence have also caused difficulty in determining the organization of this genomic region. ''SMN1'' and ''SMN2'' are nearly identical and encode the same protein. The critical sequence difference between the two is a single nucleotide in exon 7 which is thought to be an exon splice enhancer. It is thought that gene conversion events may involve the two genes, leading to varying copy numbers of each gene. Clinical significance Mutations in ''SMN1'' are associate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zygosity
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same locus (genetics), loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal Sex-determination system#Chromosomal determination, sex-determination system. If both alleles of a diploid organism are the same, the organism is #Homozygous, homozygous at that locus. If they are different, the organism is #Heterozygous, heterozygous at that locus. If one allele is missing, it is #Hemizygous, hemizygous, and, if both alleles are missing, it is #Nullizygous, nullizygous. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spinal Muscular Atrophy
Spinal muscular atrophy (SMA) is a rare neuromuscular disorder that results in the loss of motor neurons and progressive muscle wasting. It is usually diagnosed in infancy or early childhood and if left untreated it is the most common genetic cause of infant death. It may also appear later in life and then have a milder course of the disease. The common feature is the progressive weakness of voluntary muscles, with the arm, leg, and respiratory muscles being affected first. Associated problems may include poor head control, difficulties swallowing, scoliosis, and joint contractures. The age of onset and the severity of symptoms form the basis of the traditional classification of spinal muscular atrophy into a number of types. Spinal muscular atrophy is due to an abnormality (mutation) in the ''SMN1'' gene which encodes SMN, a protein necessary for the survival of motor neurons. Loss of these neurons in the spinal cord prevents signalling between the brain and skeletal mus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
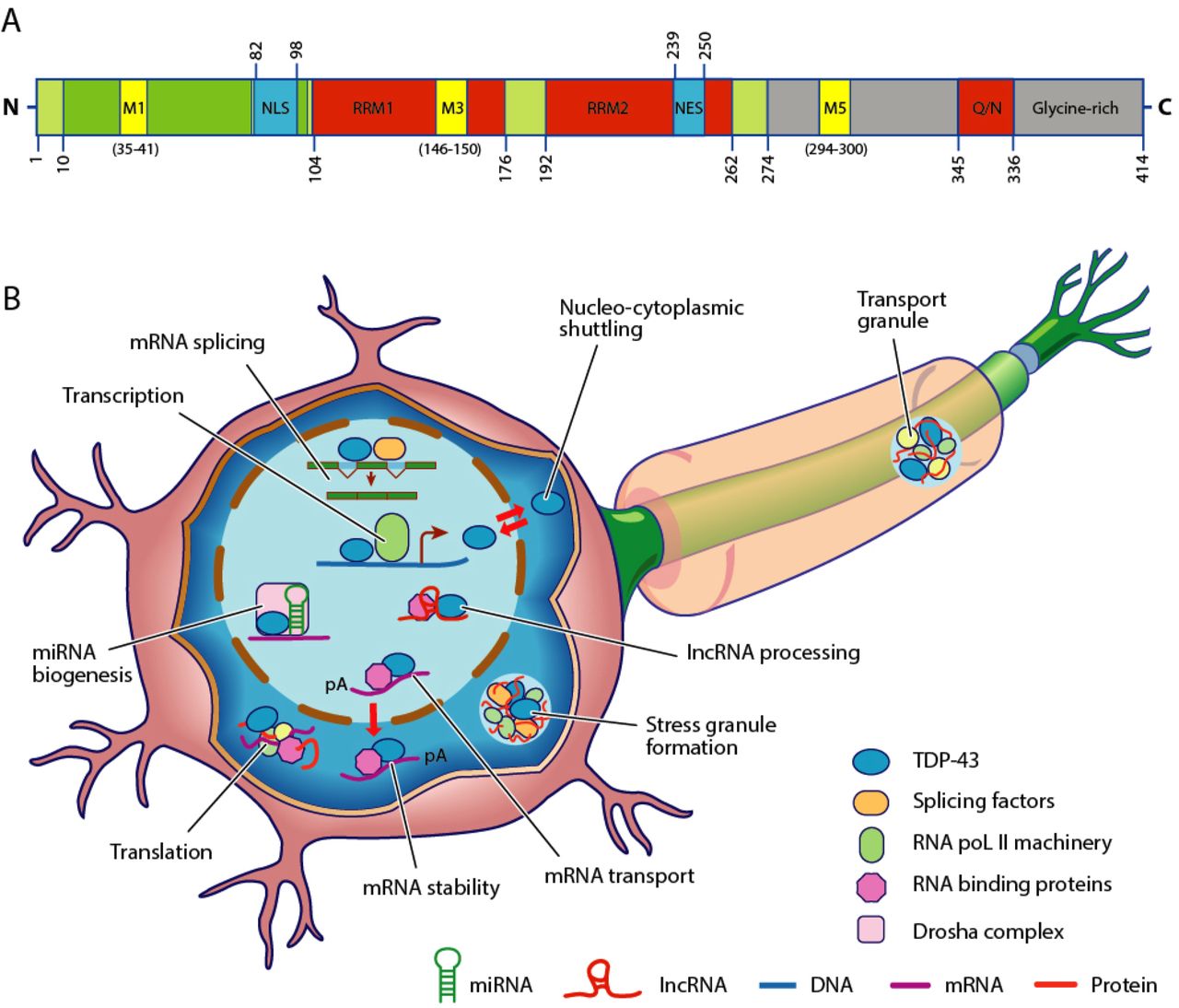
TARDBP
Transactive response DNA binding protein 43 kDa (TAR DNA-binding protein 43 or TDP-43) is a protein that in humans is encoded by the ''TARDBP'' gene. Structure TDP-43 is 414 amino acid residues long. It consists of four domains: an N-terminal domain spanning residues 1–76 (NTD) with a well-defined fold that has been shown to form a dimer or oligomer; two highly conserved folded RNA recognition motifs spanning residues 106–176 (RRM1) and 191–259 (RRM2), respectively, required to bind target RNA and DNA; an unstructured C-terminal domain encompassing residues 274–414 (CTD), which contains a glycine-rich region, is involved in protein-protein interactions, and harbors most of the mutations associated with familial amyotrophic lateral sclerosis. The entire protein devoid of large solubilising tags has been purified. The full-length protein is a dimer. The dimer is formed due to a self-interaction between two NTD domains, where the dimerisation can be propagated to fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |