|
DNA Transposon
DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I TEs, retrotransposons, that move through an RNA intermediate. DNA transposons can move in the DNA of an organism via a single-or double-stranded DNA intermediate. DNA transposons have been found in both Prokaryote, prokaryotic and Eukaryote, eukaryotic organisms. They can make up a significant portion of an organism's genome, particularly in eukaryotes. In prokaryotes, TE's can facilitate the Horizontal gene transfer, horizontal transfer of antibiotic resistance or other genes associated with virulence. After replicating and propagating in a host, all transposon copies become inactivated and are lost unless the transposon passes to a genome by starting a new life cycle with horizontal transfer. DNA transposons do not randomly insert themse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage. Mechanisms of duplication Ectopic recombination Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of it happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombina ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Soybean Rust
Soybean rust is a disease that affects soybeans and other legumes. It is caused by two types of fungi, '' Phakopsora pachyrhizi'', commonly known as Asian soybean rust, and '' Phakopsora meibomiae'', commonly known as New World soybean rust. ''P. meibomiae'' is the weaker pathogen of the two and generally does not cause widespread problems. The disease has been reported across Asia, Australia, Africa, South America and the United States. Importance Soybean is one of the most important commercial crops around the world. Asian soybean rust is the major disease that affects soybeans. It causes lesions on the leaves of soybean plants and eventually kills the plants. The disease has caused serious yield loss of soybeans. In the areas where this disease is common, the yield losses can be up to 80%. The first reported cases in the United States occurred in 2004. This population originated in northern South America or the Caribbean. The spores were most likely spread by Hurricane Ivan. I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Entamoeba
''Entamoeba'' is a genus of Amoebozoa found as internal parasites or commensals of animals. In 1875, Fedor Lösch described the first proven case of amoebic dysentery in St. Petersburg, Russia. He referred to the amoeba he observed microscopically as ''Amoeba coli''; however, it is not clear whether he was using this as a descriptive term or intended it as a formal taxonomic name. The genus ''Entamoeba'' was defined by Casagrandi and Barbagallo for the species '' Entamoeba coli'', which is known to be a commensal organism. Lösch's organism was renamed ''Entamoeba histolytica'' by Fritz Schaudinn in 1903; he later died, in 1906, from a self-inflicted infection when studying this amoeba. For a time during the first half of the 20th century the entire genus ''Entamoeba'' was transferred to '' Endamoeba'', a genus of amoebas infecting invertebrates about which little is known. This move was reversed by the International Commission on Zoological Nomenclature in the late 1950s, and '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Animal
Animals are multicellular, eukaryotic organisms in the Biology, biological Kingdom (biology), kingdom Animalia (). With few exceptions, animals heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, have myocytes and are motility, able to move, can reproduce sexually, and grow from a hollow sphere of Cell (biology), cells, the blastula, during embryonic development. Animals form a clade, meaning that they arose from a single common ancestor. Over 1.5 million extant taxon, living animal species have been species description, described, of which around 1.05 million are insects, over 85,000 are molluscs, and around 65,000 are vertebrates. It has been estimated there are as many as 7.77 million animal species on Earth. Animal body lengths range from to . They have complex ecologies and biological interaction, interactions with each other and their environments, forming intricate food webs. The scientific study of animals is known as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fungus
A fungus (: fungi , , , or ; or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and mold (fungus), molds, as well as the more familiar mushrooms. These organisms are classified as one of the kingdom (biology)#Six kingdoms (1998), traditional eukaryotic kingdoms, along with Animalia, Plantae, and either Protista or Protozoa and Chromista. A characteristic that places fungi in a different kingdom from plants, bacteria, and some protists is chitin in their cell walls. Fungi, like animals, are heterotrophs; they acquire their food by absorbing dissolved molecules, typically by secreting digestive enzymes into their environment. Fungi do not photosynthesize. Growth is their means of motility, mobility, except for spores (a few of which are flagellated), which may travel through the air or water. Fungi are the principal decomposers in ecological systems. These and other differences place fungi in a single group of related o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
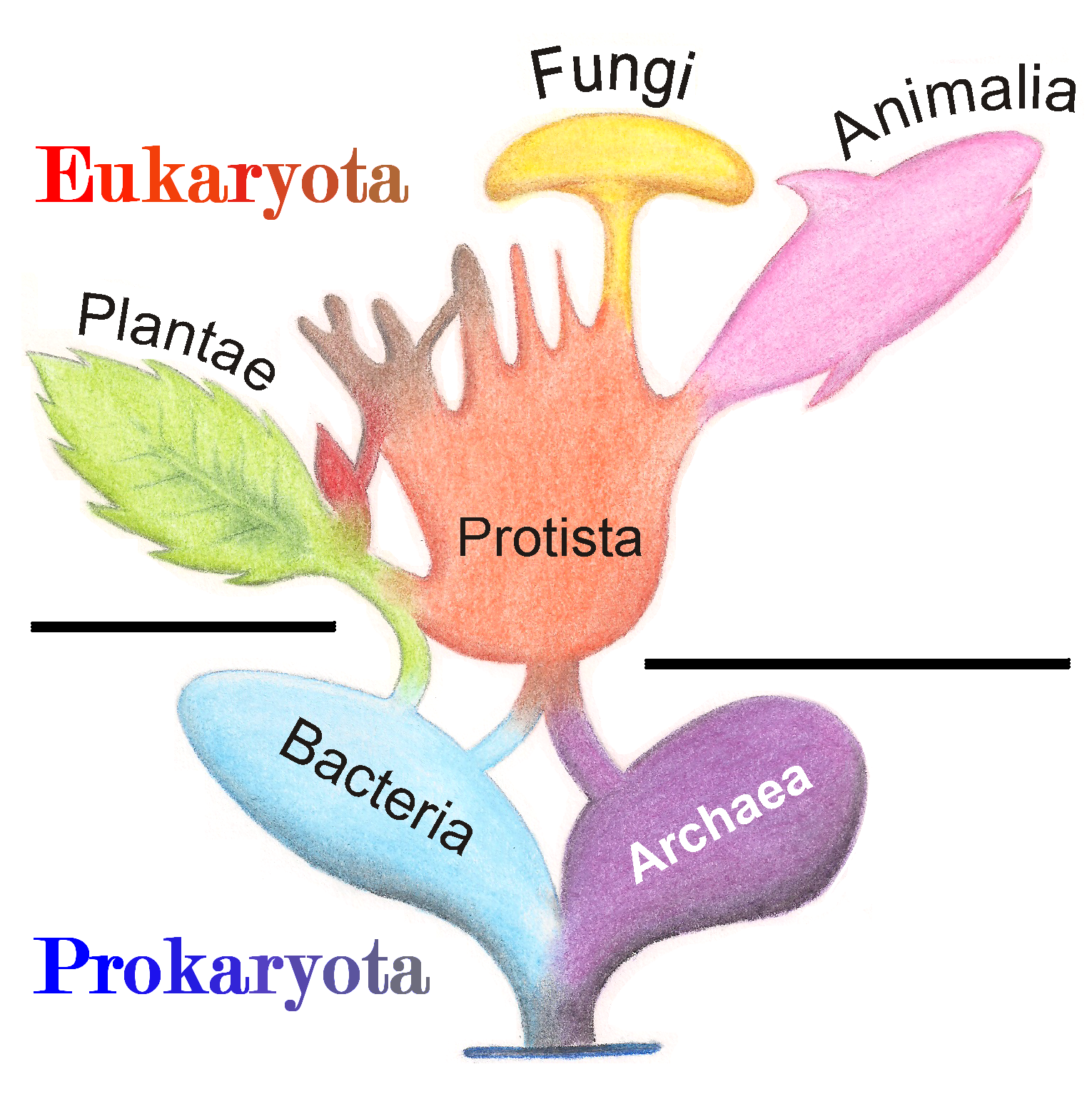
Protist
A protist ( ) or protoctist is any eukaryotic organism that is not an animal, land plant, or fungus. Protists do not form a natural group, or clade, but are a paraphyletic grouping of all descendants of the last eukaryotic common ancestor excluding land plants, animals, and fungi. Protists were historically regarded as a separate taxonomic kingdom known as Protista or Protoctista. With the advent of phylogenetic analysis and electron microscopy studies, the use of Protista as a formal taxon was gradually abandoned. In modern classifications, protists are spread across several eukaryotic clades called supergroups, such as Archaeplastida ( photoautotrophs that includes land plants), SAR, Obazoa (which includes fungi and animals), Amoebozoa and " Excavata". Protists represent an extremely large genetic and ecological diversity in all environments, including extreme habitats. Their diversity, larger than for all other eukaryotes, has only been discovered in rece ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposons
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome. The discovery of mobile genetic elements earned Barbara McClintock a Nobel Prize in 1983. There are at least two classes of TEs: Class I TEs or retrotransposons generally function via reverse transcription, while Class II TEs or DNA transposons encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. Discovery by Barbara McClintock Barbara McClintock discovered the first TEs in maize (''Zea mays'') at the Cold Spring Harbor Laboratory in New York. McClintock was experimenting with maize plants that had broken chromosomes. In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (Style (botany), style) of the flower received pollen from its own anther. These kernels came fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
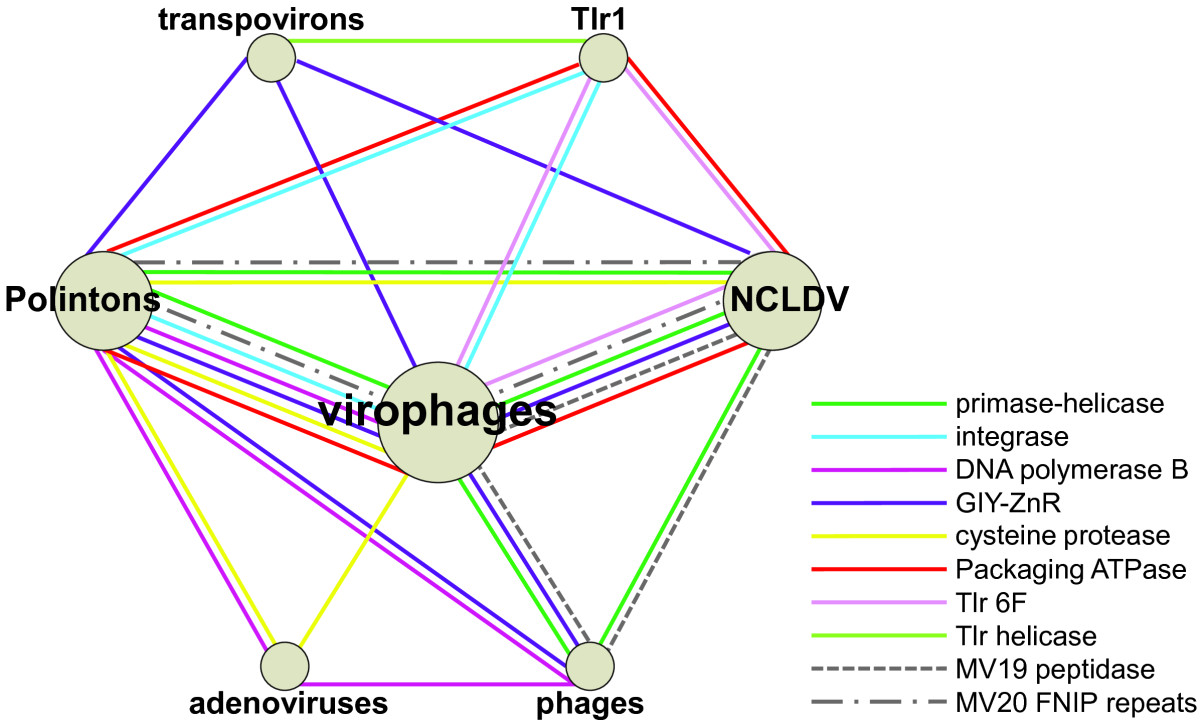
Polinton
Polintons (also called Mavericks) are large DNA transposons which contain genes with homology (biology), homology to virus, viral proteins and which are often found in eukaryotic genomes. They were first discovered in the mid-2000s and are the largest and most complex known DNA transposons. Polintons encode up to 10 individual proteins and derive their name from two key proteins, a DNA polymerase, DNA polymerase and a retroviral-like integrase, integrase. Properties A typical polinton is around 15–20 kilobase pairs in size, though examples have been described up to 40kb. Polintons encode up to 10 proteins, the key elements being the protein-primed type B DNA polymerase and the retroviral-like integrase from which they derive their name. Polintons are sometimes referred to as "self-synthesizing" transposons, because they encode the proteins necessary to replicate themselves. Most polintons also encode an adenoviral-like cysteine protease, an FtsK-like ATPase, and proteins with homol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polinton Transposition Mechanism
Polintons (also called Mavericks) are large DNA transposons which contain genes with homology to viral proteins and which are often found in eukaryotic genomes. They were first discovered in the mid-2000s and are the largest and most complex known DNA transposons. Polintons encode up to 10 individual proteins and derive their name from two key proteins, a DNA polymerase and a retroviral-like integrase. Properties A typical polinton is around 15–20 kilobase pairs in size, though examples have been described up to 40kb. Polintons encode up to 10 proteins, the key elements being the protein-primed type B DNA polymerase and the retroviral-like integrase from which they derive their name. Polintons are sometimes referred to as "self-synthesizing" transposons, because they encode the proteins necessary to replicate themselves. Most polintons also encode an adenoviral-like cysteine protease, an FtsK-like ATPase, and proteins with homology to the jelly-roll fold structure of vir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized nucleic acid strands (hence '' helic- + -ase''), via the energy gained from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing viral nucleic acids during infection and fulfill an immunological function. Genetic mutations that affect helicase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called '' restriction endonucleases'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |