|
Reductase
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NADH, NAD+ as Cofactor (biochemistry), cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, Photosystem, chloroplasts and mitochondria, including respiratory complexes Respiratory complex I, I, complex II, II and complex III, III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix. in Membranome database Reactions For example, an enzyme that ...[...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Respiratory Complex I
Respiratory complex I, (also known as NADH:ubiquinone oxidoreductase, Type I NADH dehydrogenase and mitochondrial complex I) is the first large protein complex of the Electron transport chain, respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria. This enzyme is essential for the normal functioning of cells, and mutations in its subunits lead to a wide range of inherited neuromuscular and metabolic disorders. Defects in this enzyme are responsible for the development of several pathological processes such as Reperfusion injury, ischemia/reperfusion damage (stroke and Myocardial infarction, cardiac infarction), Parkinson's disease and others. Function Complex I is the first enzyme of the Electron transport chain#Mitochondrial electron transport chains, mitochondrial electron tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADH
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD and NADH (H for hydrogen), respectively. In cellular metabolism, NAD is involved in redox reactions, carrying electrons from one reaction to another, so it is found in two forms: NAD is an oxidizing agent, accepting electrons from other molecules and becoming reduced; with H+, this reaction forms NADH, which can be used as a reducing agent to donate electrons. These electron transfer reactions are the main function of NAD. It is also used in other cellular processes, most notably as a substrate of enzymes in adding or removing chemical groups to or from proteins, in posttranslational modifications. Because of the import ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Transport Chain
An electron transport chain (ETC) is a series of protein complexes and other molecules which transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. Many of the enzymes in the electron transport chain are embedded within the membrane. The flow of electrons through the electron transport chain is an exergonic process. The energy from the redox reactions creates an electrochemical proton gradient that drives the synthesis of adenosine triphosphate (ATP). In aerobic respiration, the flow of electrons terminates with molecular oxygen as the final electron acceptor. In anaerobic respiration, other electron acceptors are used, such as sulfate. In an electron transport chain, the redox reactions are driven by the difference in the Gibbs free energy of reactants and products. The free energy released when ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dehydrogenase
A dehydrogenase is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example, alcohol dehydrogenase catalyzes the oxidation of ethanol to acetaldehyde in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde. IUBMB classification Oxidoreductases, enzymes that catalyze oxidation-reduction reactions, constitute Class EC 1 of the IUBMB classification of enzyme-catalyzed reactions. Any of these may be called dehydrogenases, especially those in which NAD+ is the electron acceptor (oxidant), but reductase is also used when the physiological emphasis on reduction of the substrate, and oxidase is used ''only'' when O2 is the electron acceptor. The systematic name of an oxidoreductase is "donor:acceptor oxidore ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be classified into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a " prosthetic group", which consists of a coenzyme that is tightly (or even covalently and, therefore, permanently) bound to a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complex II
Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and oxidative phosphorylation. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential. In step 6 of the citric acid cycle, SQR catalyzes the oxidation of succinate to fumarate with the reduction of ubiquinone to ubiquinol. This occurs in the inner mitochondrial membrane by coupling the two reactions together. Structure Subunits Mitochondrial and many bacterial SQRs are composed of four structurally different subunits: two hydrophilic and two hydrophobic. The first two subunits, a flavoprotein (SDHA) and an iron-sulfur protein (SDHB), form a hydrophilic head where enzymatic activity of the complex takes place. SD ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alcohol Oxidoreductases
Alcohol oxidoreductases are oxidoreductase enzymes that act upon an alcohol functional group In organic chemistry, a functional group is any substituent or moiety (chemistry), moiety in a molecule that causes the molecule's characteristic chemical reactions. The same functional group will undergo the same or similar chemical reactions r .... They are classified under "1.1" in the EC number numbering system. References External links * EC 1.1 {{1.1-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADP+
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form, whereas NADP is the oxidized form. NADP is used by all forms of cellular life. NADP is essential for life because it is needed for cellular respiration. NADP differs from NAD by the presence of an additional phosphate group on the 2' position of the ribose ring that carries the adenine moiety. This extra phosphate is added by NAD+ kinase and removed by NADP+ phosphatase. Biosynthesis NADP In general, NADP+ is synthesized before NADPH is. Such a reaction usually starts with NAD+ from either the de-novo or the salvage pathway, with NAD+ kinase adding the extra phosphate group. ADP-ribosyl cyclase allows for synthesis from nicotinamide in the salvage pathway, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanol Dehydrogenase
In enzymology, a methanol dehydrogenase (MDH) is an enzyme that catalyzes the chemical reaction: : CH3OH \rightleftharpoons CH2O + 2 electrons + 2H+ How the electrons are captured and transported depends upon the kind of methanol dehydrogenase. There are three main types of MDHs: Nicotinamide adenine dinucleotide, NAD+-dependent MDH, Pyrroloquinoline quinone, pyrrolo-quinoline quinone dependent MDH, and oxygen-dependent alcohol oxidase. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The List of enzymes, systematic name of this enzyme class is methanol:NAD+ oxidoreductase. This enzyme participates in methane metabolism. Classes of Methanol Dehydrogenase NAD+ Dependent MDH A common electron acceptor in biological systems is nicotinamide adenine dinucleotide (NAD+); some enzymes use a related molecule called nicotinamide adenine dinucleotide phosphate (NADP+). An NAD+-dependent methano ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidase
In biochemistry, an oxidase is an oxidoreductase (any enzyme that catalyzes a redox reaction) that uses dioxygen (O2) as the electron acceptor. In reactions involving donation of a hydrogen atom, oxygen is reduced to water (H2O) or hydrogen peroxide (H2O2). Some oxidation reactions, such as those involving monoamine oxidase or xanthine oxidase, typically do not involve free molecular oxygen. The oxidases are a subclass of the oxidoreductases. The use of dioxygen is the only unifying feature; in the EC classification, these enzymes are scattered in many categories. Examples An important example is EC 7.1.1.9 cytochrome c oxidase, the key enzyme that allows the body to employ oxygen in the generation of energy and the final component of the electron transfer chain. Other examples are: * EC 1.1.3.4 Glucose oxidase * EC 1.4.3.4 Monoamine oxidase * EC 1.14.-.- Cytochrome P450 oxidase * EC 1.6.3.1 NADPH oxidase * EC 1.17.3.2 Xanthine oxidase * EC 1.1.3.8 L-gulonolactone oxidase * EC 1. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
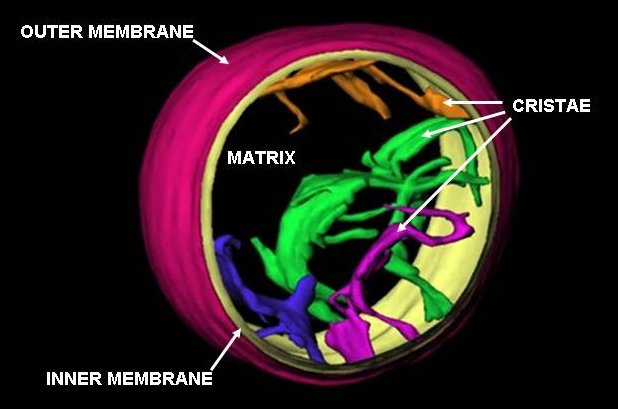
Mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal '' Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number of unicellular organisms, such as microspo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |