|
Centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase. There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency. Any piece of DNA with the point centromere DNA sequence on it will typically form a centr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Telocentric
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase. There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency. Any piece of DNA with the point centromere DNA sequence on it will typically form a centromere if p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Kinetochore
A kinetochore (, ) is a flared oblique-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers, which can be thought of as the ropes pulling chromosomes apart, attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term ''kinetochore'' (= movement place) has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932. Monocentric organisms, including vertebrates, fungi, and most plants, have a single centromeric region on each chromosome which assembles a single, localized kinetochore. Holocentric organisms, such as nematodes a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
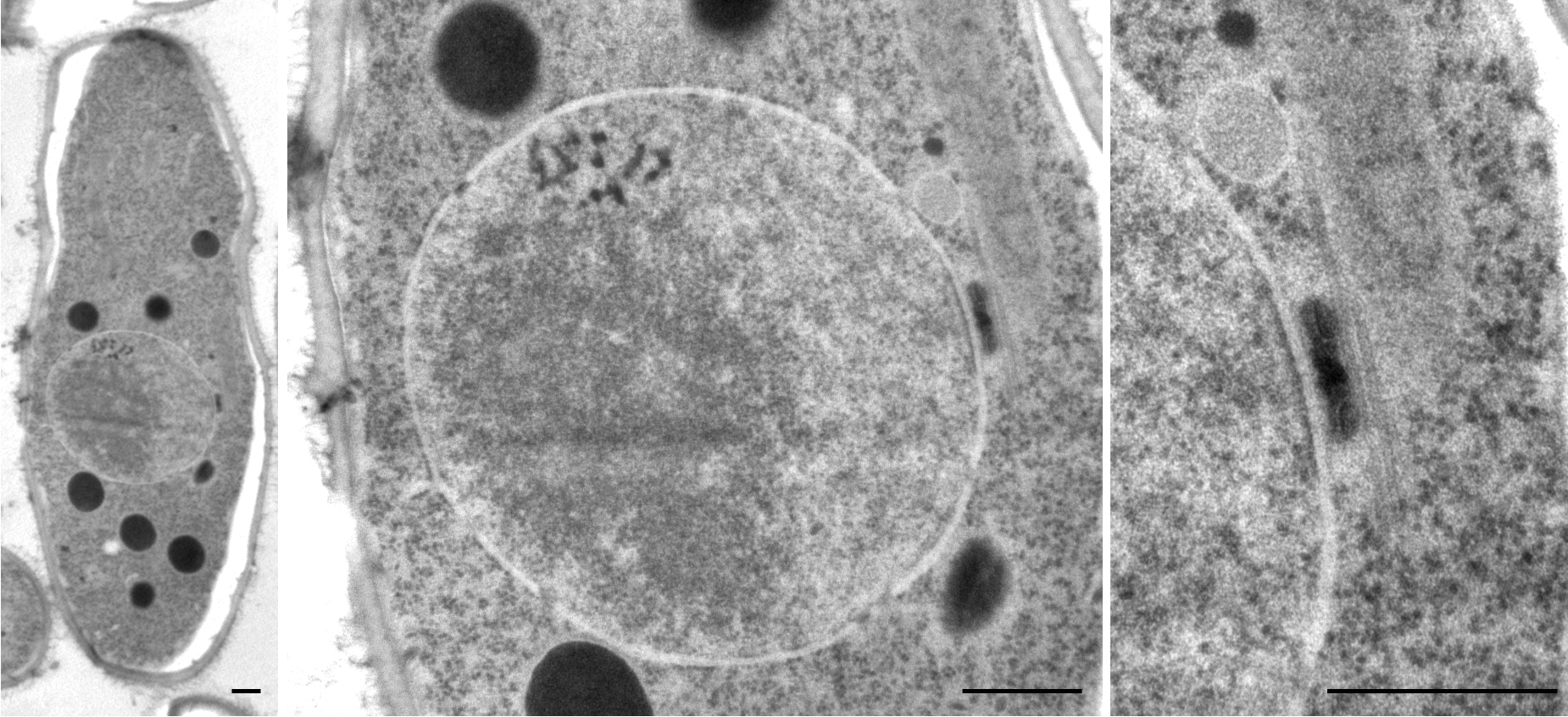
Schizosaccharomyces Pombe
''Schizosaccharomyces pombe'', also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs. These cells maintain their shape by growing exclusively through the cell tips and divide by medial fission to produce two daughter cells of equal size, which makes them a powerful tool in cell cycle research. Fission yeast was isolated in 1893 by Paul Lindner from East African millet beer. The species name ''pombe'' is the Swahili word for beer. It was first developed as an experimental model in the 1950s: by Urs Leupold for studying genetics, and by Murdoch Mitchison for studying the cell cycle. Paul Nurse, a fission ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most important of these proteins are the histones. Aided by chaperone proteins, the histones bind to and condense the DNA molecule to maintain its integrity. These eukaryotic chromosomes display a complex three-dimensional structure that has a significant role in transcriptional regulation. Normally, chromosomes are visible under a light microscope only during the metaphase of cell division, where all chromosomes are aligned in the center of the cell in their condensed form. Before this stage occurs, each chromosome is duplicated ( S phase), and the two copies are joined by a centromere—resulting in either an X-shaped structure if the centromere is located equatorially, or a two-armed structure if the centromere is located distally; the jo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Karyotype
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities. A karyogram or idiogram is a graphical depiction of a karyotype, wherein chromosomes are generally organized in pairs, ordered by size and position of centromere for chromosomes of the same size. Karyotyping generally combines light microscopy and photography in the metaphase of the cell cycle, and results in a photomicrographic (or simply micrographic) karyogram. In contrast, a schematic karyogram is a designed graphic representation of a karyotype. In schematic karyograms, just one of the sister chromatids of each chromosome is generally shown for brevity, and in reality they are generally so close together that they look as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identical cells in which the total number of chromosomes is maintained. Mitosis is preceded by the S phase of interphase (during which DNA replication occurs) and is followed by telophase and cytokinesis, which divide the cytoplasm, organelles, and cell membrane of one cell into two new cell (biology), cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic phase (M phase) of a cell cycle—the cell division, division of the mother cell into two daughter cells genetically identical to each other. The process of mitosis is divided into stages corresponding to the completion of one set of activities and the start of the next. These stages are preprophase (specific to plant ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Cohesin
Cohesin is a protein complex that mediates Establishment of sister chromatid cohesion, sister chromatid cohesion, homologous recombination, and Topologically associating domain, DNA looping. Cohesin is formed of SMC3, SMC1A, SMC1, RAD21, SCC1 and SCC3 (STAG1, SA1 or STAG2, SA2 in humans). Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC proteins, SMC family of protein complexes which includes Condensin, Nucleoid#MukBEF, MukBEF and SMC-ScpAB. Cohesin was separately discovered in budding yeast (''Saccharomyces cerevisiae'') both by Douglas Koshland and Kim Nasmyth in 1997. Structure and subunits Cohesin is a multi-subunit protein complex, made up of SMC1, SMC3, RAD21 and SCC3 (SA1 or SA2). SMC1 and SMC3 are members of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |