|
CRISPR Gene Editing
CRISPR gene editing (; pronounced like "crisper"; an abbreviation for "clustered regularly interspaced short palindromic repeats") is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed or new ones added ''in vivo''. The technique is considered highly significant in biotechnology and medicine as it enables editing genomes ''in vivo'' and is precise, cost-effective, and efficient. It can be used in the creation of new medicines, agriculture, agricultural products, and genetically modified organisms, or as a means of controlling pathogens and pest control, pests. It also offers potential in the treatment of inherited genetic diseases as well as diseases arisi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Jennifer Doudna
Jennifer Anne Doudna (; born February 19, 1964) is an American biochemist who has pioneered work in CRISPR gene editing, and made other fundamental contributions in biochemistry and genetics. She received the 2020 Nobel Prize in Chemistry, with Emmanuelle Charpentier, "for the development of a method for genome editing." She is the Li Ka-shing, Li Ka Shing Chancellor, Chancellor's Professor, Chair Professor in the department of chemistry and the department of molecular and cell biology at the University of California, Berkeley. She has been an investigator with the Howard Hughes Medical Institute since 1997. In 2012, Doudna and Emmanuelle Charpentier were the first to propose that CRISPR-Cas9 (enzymes from bacteria that control microbial immunity) could be used for programmable editing of genomes, which has been called one of the most significant discoveries in the history of biology. Since then, Doudna has been a leading figure in what is referred to as the "CRISPR revolution" ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
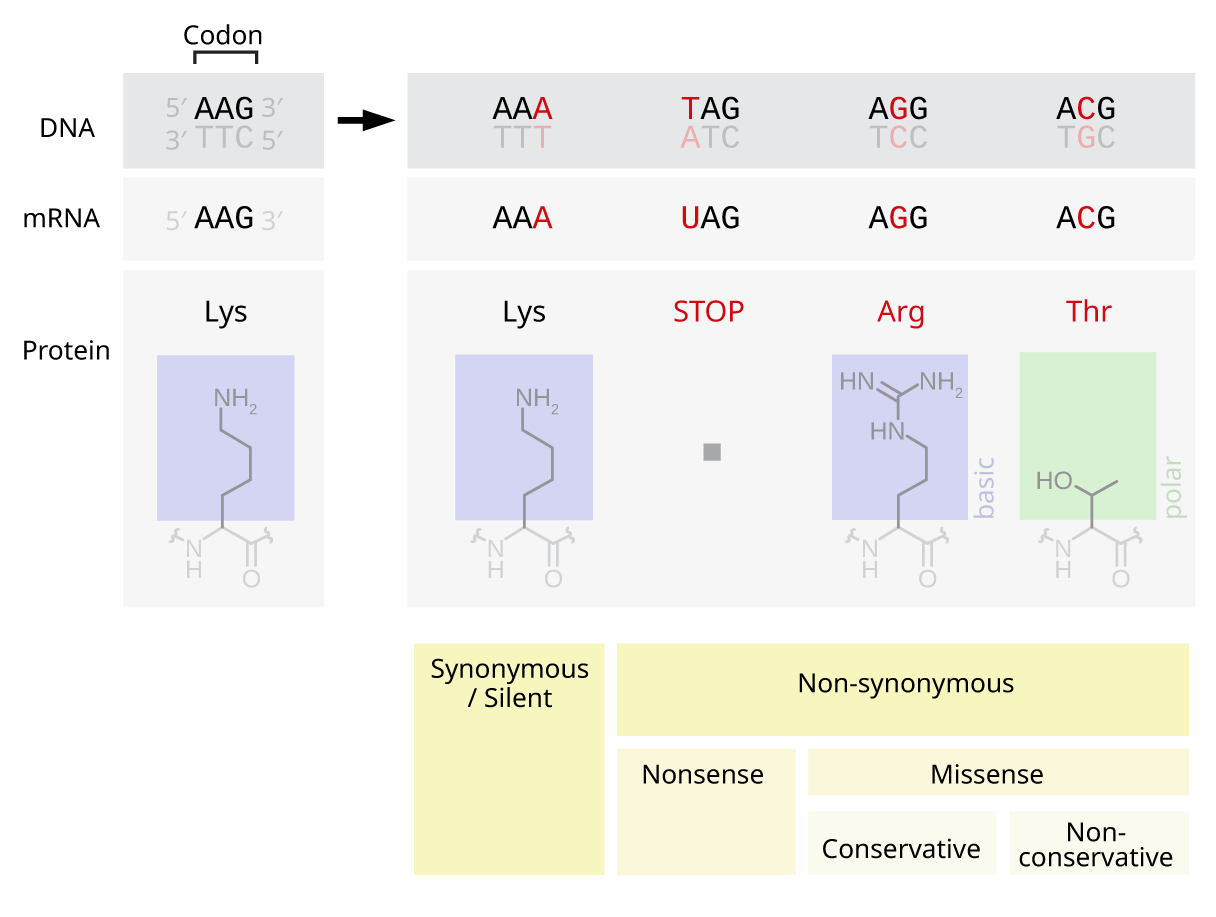
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. Synonymous substitution, synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either Transcription (genetics), transcription or Translation (biology), translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders. Gene silencing is often considered the same as gene knockdown. When genes are silenced, their expression is reduced. In contrast, when genes are knocked out, they are completely erased from the organism's genome and, thus, have no expression. Gene silencing is considered a gene knockdown mechanism since the methods used to silence genes, such as RNAi, CRISPR, or siRNA, generally reduce the expression of a gene by at least 70% but do not eliminate it. Methods using gene silencing are often considered better than gene knockouts since they allo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptococcus Pyogenes
''Streptococcus pyogenes'' is a species of Gram-positive, aerotolerant bacteria in the genus '' Streptococcus''. These bacteria are extracellular, and made up of non-motile and non-sporing cocci (round cells) that tend to link in chains. They are clinically important for humans, as they are an infrequent, but usually pathogenic, part of the skin microbiota that can cause group A streptococcal infection. ''S. pyogenes'' is the predominant species harboring the Lancefield group A antigen, and is often called group A ''Streptococcus'' (GAS). However, both '' Streptococcus dysgalactiae'' and the '' Streptococcus anginosus'' group can possess group A antigen as well. Group A streptococci, when grown on blood agar, typically produce small (2–3 mm) zones of beta-hemolysis, a complete destruction of red blood cells. The name group A (beta-hemolytic) ''Streptococcus'' is thus also used. The species name is derived from Greek words meaning 'a chain' () of berries ( atiniz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of Outline of life forms, life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal Kingdom (biology), kingdom Asgard (Archaea), Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only Two-domain system, two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as Flagellated cell, flagellated cells. The leading evolutiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Editing
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly insert genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases (FokI and CRISPR associated protein, Cas), and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ). History Genome editing was pioneered in the 1990s, before the advent of the common current nuclease-based gene-editing platforms, but its use was limited by low efficiencies of editing. Genome editing with engineered nucleases, i.e. all three m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
POLQ
DNA polymerase theta is an enzyme that in humans is encoded by the ''POLQ'' gene. This polymerase plays a key role in one of the three major double strand break repair pathways: theta-mediated end joining (TMEJ). Most double-strand breaks are repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR). However, in some contexts, NHEJ and HR are insufficient and TMEJ is the only solution to repair the break. TMEJ is often described as alternative NHEJ, but differs in that it lacks a requirement for the Ku heterodimer, and it can only act on resected DNA ends. Following annealing of short (i.e., a few nucleotides) regions on the DNA overhangs, DNA polymerase theta catalyzes template-dependent DNA synthesis across the broken ends, stabilizing the paired structure. Polymerase theta's mutational signature TMEJ is intrinsically mutagenic, since polymerase theta uses homologous nucleotides from both break ends to initiate repair, which leads to loss of one set of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology Directed Repair
Homology-directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining ( NHEJ) takes place instead. Cancer suppression HDR is important for suppressing the formation of cancer. HDR maintains genomic stability by repairing broken DNA strands; it is assumed to be error free because of the use of a template. When a double strand DNA lesion is repaired by NHEJ there is no validating DNA template present so it may result in a novel DNA strand formation with loss of information. A different nucleotide sequence in the DNA strand results in a different protein e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virginijus Šikšnys
Virginijus Šikšnys (born 26 January 1956) is a Lithuanian biochemist and a professor at Vilnius University. He is a chief scientist at the Vilnius University Institute of Biotechnology. Biography Šikšnys studied organic chemistry at Vilnius University, receiving his Bachelor's degree in 1978. He then moved to Lomonosov Moscow State University, where he studied enzyme kinetics and received the Candidate of Sciences degree (equivalent to PhD at that time) in 1983. His PhD advisor was Karel Martinek from, at that time, Czechoslovakia. From 1982 till 1993 he worked at the Institute of Applied Enzymology in Vilnius. In 1993 he was a visiting scientist in Robert Huber’s laboratory at the Max-Planck-Institut für Biochemie, Martinsried, Germany. Since 1995 Šikšnys is the chief scientist and head of the Department of Protein-DNA Interactions at the Vilnius University Institute of Biotechnology, since 2006 – professor at Vilnius University and a member of the Lithuania ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |