|
Binding Site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins (resulting in a protein-protein interaction), enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible (transient and non-covalent), but can also be covalent reversible or irreversible. Function Binding of a ligand to a binding site on protein often triggers a change in conformation in the protein and results in altered cellular function. Hence binding site on protein are critical parts of signal transduction pathways. Types of ligands include neurotransmitters, toxins, neuropeptides, and steroid hormones. Binding sites incur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glucose Binding Hexokinase
Glucose is a simple sugar with the Chemical formula#Molecular formula, molecular formula . Glucose is overall the most abundant monosaccharide, a subcategory of carbohydrates. Glucose is mainly made by plants and most algae during photosynthesis from water and carbon dioxide, using energy from sunlight, where it is used to make cellulose in cell walls, the most abundant carbohydrate in the world. In energy metabolism, glucose is the most important source of energy in all organisms. Glucose for metabolism is stored as a polymer, in plants mainly as starch and amylopectin, and in animals as glycogen. Glucose circulates in the blood of animals as blood sugar. The naturally occurring form of glucose is -glucose, while L-glucose, -glucose is produced synthetically in comparatively small amounts and is less biologically active. Glucose is a monosaccharide containing six carbon atoms and an Aldehyde , aldehyde group, and is therefore an aldohexose. The glucose molecule can exist in an op ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Steroid Hormone
A steroid hormone is a steroid that acts as a hormone. Steroid hormones can be grouped into two classes: corticosteroids (typically made in the adrenal cortex, hence ''cortico-'') and sex steroids (typically made in the gonads or placenta). Within those two classes are five types according to the receptors to which they bind: glucocorticoids and mineralocorticoids (both corticosteroids) and androgens, estrogens, and progestogens (sex steroids). Vitamin D derivatives are a sixth closely related hormone system with homologous receptors. They have some of the characteristics of true steroids as receptor ligands. Steroid hormones help control metabolism, inflammation, immune functions, salt and water balance, development of sexual characteristics, and the ability to withstand injury and illness. The term steroid describes both hormones produced by the body and artificially produced medications that duplicate the action for the naturally occurring steroids. Synthesis The natur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Support-vector Machine
In machine learning, support vector machines (SVMs, also support vector networks) are supervised learning models with associated learning algorithms that analyze data for classification and regression analysis. Developed at AT&T Bell Laboratories by Vladimir Vapnik with colleagues (Boser et al., 1992, Guyon et al., 1993, Cortes and Vapnik, 1995, Vapnik et al., 1997) SVMs are one of the most robust prediction methods, being based on statistical learning frameworks or VC theory proposed by Vapnik (1982, 1995) and Chervonenkis (1974). Given a set of training examples, each marked as belonging to one of two categories, an SVM training algorithm builds a model that assigns new examples to one category or the other, making it a non-probabilistic binary linear classifier (although methods such as Platt scaling exist to use SVM in a probabilistic classification setting). SVM maps training examples to points in space so as to maximise the width of the gap between the two categories. New ex ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Druggability
Druggability is a term used in drug discovery to describe a biological target (such as a protein) that is known to or is predicted to bind with high affinity to a drug. Furthermore, by definition, the binding of the drug to a druggable target must alter the function of the target with a therapeutic benefit to the patient. The concept of druggability is most often restricted to small molecules (low molecular weight organic substances) but also has been extended to include biologic medical products such as therapeutic monoclonal antibodies. Drug discovery comprises a number of stages that lead from a biological hypothesis to an approved drug. Target identification is typically the starting point of the modern drug discovery process. Candidate targets may be selected based on a variety of experimental criteria. These criteria may include disease linkage (mutations in the protein are known to cause a disease), mechanistic rationale (for example, the protein is part of a regulatory ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catabolism
Catabolism () is the set of metabolic pathways that breaks down molecules into smaller units that are either oxidized to release energy or used in other anabolic reactions. Catabolism breaks down large molecules (such as polysaccharides, lipids, nucleic acids, and proteins) into smaller units (such as monosaccharides, fatty acids, nucleotides, and amino acids, respectively). Catabolism is the breaking-down aspect of metabolism, whereas anabolism is the building-up aspect. Cells use the monomers released from breaking down polymers to either construct new polymer molecules or degrade the monomers further to simple waste products, releasing energy. Cellular wastes include lactic acid, acetic acid, carbon dioxide, ammonia, and urea. The formation of these wastes is usually an oxidation process involving a release of chemical free energy, some of which is lost as heat, but the rest of which is used to drive the synthesis of adenosine triphosphate (ATP). This molecule ac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphofructokinase
Phosphofructokinase (PFK) is a kinase enzyme that phosphorylates fructose 6-phosphate in glycolysis. Function The enzyme-catalysed transfer of a phosphoryl group from ATP is an important reaction in a wide variety of biological processes. Phosphofructokinase catalyses the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate, a key regulatory step in the glycolytic pathway. It is allosterically inhibited by ATP and allosterically activated by AMP, thus indicating the cell's energetic needs when it undergoes the glycolytic pathway. PFK exists as a homotetramer in bacteria and mammals (where each monomer possesses 2 similar domains) and as an octomer in yeast (where there are 4 alpha- (PFK1) and 4 beta-chains (PFK2), the latter, like the mammalian monomers, possessing 2 similar domains). This protein may use the morpheein model of allosteric regulation. PFK is about 300 amino acids in length, and structural studies of the bacterial enzyme have shown it compr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allosteric Regulation
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. Allosteric regulation is also particularly important in the cell's ability to adjust enzyme activity. The term ''allostery'' comes from the Ancient Greek ''allos'' (), "other", and ''stereos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Inhibition
An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction. An enzyme inhibitor stops ("inhibits") this process, either by binding to the enzyme's active site (thus preventing the substrate itself from binding) or by binding to another site on the enzyme such that the enzyme's catalysis of the reaction is blocked. Enzyme inhibitors may bind reversibly or irreversibly. Irreversible inhibitors form a chemical bond with the enzyme such that the enzyme is inhibited until the chemical bond is broken. By contrast, reversible inhibitors bind non-covalently and may spontaneously leave the enzyme, allowing the enzyme to resume its function. Reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heme
Heme, or haem (pronounced / hi:m/ ), is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver. In biochemical terms, heme is a coordination complex "consisting of an iron ion coordinated to a porphyrin acting as a tetradentate ligand, and to one or two axial ligands." The definition is loose, and many depictions omit the axial ligands. Among the metalloporphyrins deployed by metalloproteins as prosthetic groups, heme is one of the most widely used and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigment in blood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and endothelial nitric oxide synthase. The word ''haem'' is derived from Greek ''haima'' meaning "blood". Function Hemoproteins have diverse biological fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
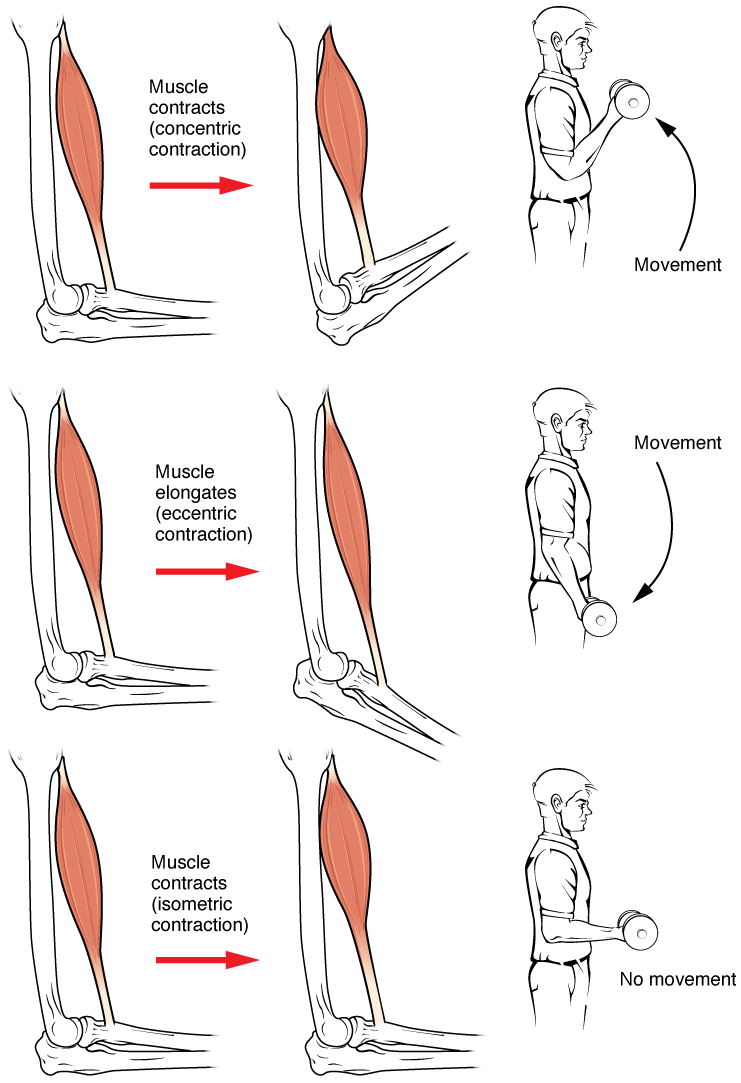
Muscle Contraction
Muscle contraction is the activation of tension-generating sites within muscle cells. In physiology, muscle contraction does not necessarily mean muscle shortening because muscle tension can be produced without changes in muscle length, such as when holding something heavy in the same position. The termination of muscle contraction is followed by muscle relaxation, which is a return of the muscle fibers to their low tension-generating state. For the contractions to happen, the muscle cells must rely on the interaction of two types of filaments which are the thin and thick filaments. Thin filaments are two strands of actin coiled around each, and thick filaments consist of mostly elongated proteins called myosin. Together, these two filaments form myofibrils which are important organelles in the skeletal muscle system. Muscle contraction can also be described based on two variables: length and tension. A muscle contraction is described as isometric if the muscle tension change ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sliding Filament Theory
The sliding filament theory explains the mechanism of muscle contraction based on muscle proteins that slide past each other to generate movement. According to the sliding filament theory, the myosin ( thick filaments) of muscle fibers slide past the actin ( thin filaments) during muscle contraction, while the two groups of filaments remain at relatively constant length. The theory was independently introduced in 1954 by two research teams, one consisting of Andrew Huxley and Rolf Niedergerke from the University of Cambridge, and the other consisting of Hugh Huxley and Jean Hanson from the Massachusetts Institute of Technology. It was originally conceived by Hugh Huxley in 1953. Andrew Huxley and Niedergerke introduced it as a "very attractive" hypothesis. Before the 1950s there were several competing theories on muscle contraction, including electrical attraction, protein folding, and protein modification. The novel theory directly introduced a new concept called cross-bridge t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uncompetitive Inhibitor
Uncompetitive inhibition, also known as anti-competitive inhibition, takes place when an enzyme inhibitor binds only to the complex formed between the enzyme and the substrate (the E-S complex). Uncompetitive inhibition typically occurs in reactions with two or more substrates or products. While uncompetitive inhibition requires that an enzyme-substrate complex must be formed, non-competitive inhibition can occur with or without the substrate present. Uncompetitive inhibition is distinguished from competitive inhibition by two observations: first uncompetitive inhibition cannot be reversed by increasing and second, as shown, the Lineweaver–Burk plot yields parallel rather than intersecting lines. This behavior is found in the inhibition of acetylcholinesterase by tertiary amines (R3N). Such compounds bind to the enzyme in its various forms, but the acyl-intermediate-amine complex cannot break down into enzyme plus product. Mechanism As inhibitor binds, the amount of ES comp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |