Synteny on:
[Wikipedia]
[Google]
[Amazon]
 In
In
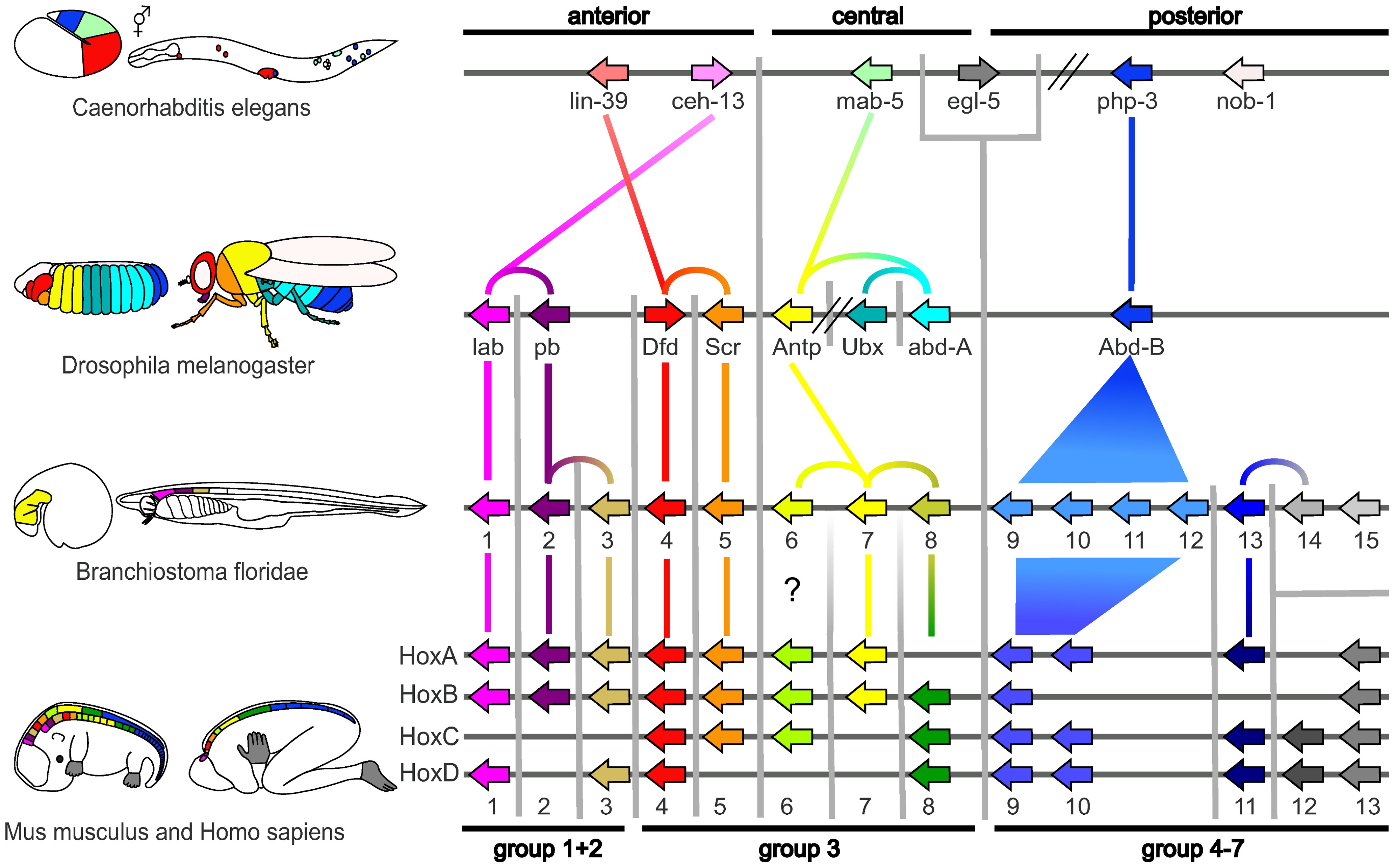 The term is currently (since ~2000) more commonly used to describe preservation of the precise order of genes on a chromosome passed down from a common ancestor, despite more "old school" geneticists rejecting what they perceive as a misappopriation of the term, preferring ''collinearity'' instead.
The analysis of synteny in the gene order sense has several applications in genomics. Shared synteny is one of the most reliable criteria for establishing the orthology of genomic regions in different species. Additionally, exceptional conservation of synteny can reflect important functional relationships between genes. For example, the order of genes in the "
The term is currently (since ~2000) more commonly used to describe preservation of the precise order of genes on a chromosome passed down from a common ancestor, despite more "old school" geneticists rejecting what they perceive as a misappopriation of the term, preferring ''collinearity'' instead.
The analysis of synteny in the gene order sense has several applications in genomics. Shared synteny is one of the most reliable criteria for establishing the orthology of genomic regions in different species. Additionally, exceptional conservation of synteny can reflect important functional relationships between genes. For example, the order of genes in the "
ACT (Artemis Comparison Tool)
— Probably the most used synteny software program used in comparative genomics.
Comparative Maps
NIH's National Library of Medicine NCBI link to Gene Homology resources, and Comparative Chromosome Maps of the Human, Mouse, and Rat.
More information on synteny and its use in comparative cereal genomics.
NCBI Home Page
NIH's National Library of Medicine NCBI (National Center for Biotechnology Information) link to a tremendous number of resources.
SimpleSynteny
A free browser-based tool to compare and visualize microsynteny across multiple genomes for a set of genes.
Synteny server
Server for Synteny Identification and Analysis of Genome Rearrangement—the Identification of synteny and calculating reversal distances.
PlantSyntenyViewer
A web based visualisation tool allowing to navigate on genomes and visualizing the Synteny conservation among several datasets (cereals, dicotyledons, animals, a Wheat-based one...) Bioinformatics Classical genetics
 In
In genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinian friar wor ...
, the term synteny refers to two related concepts:
* In classical genetics, ''synteny'' describes the physical co-localization of genetic loci
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total ...
on the same chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
within an individual or species
In biology, a species is the basic unit of classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate s ...
.
* In current biology, ''synteny'' more commonly refers to ''colinearity'', i.e. conservation of blocks of order within two sets of chromosomes that are being compared with each other. These blocks are referred to as ''syntenic blocks''.
The Encyclopædia Britannica gives the following description of synteny, using the modern definition:
Etymology
''Synteny'' is aneologism
A neologism Greek νέο- ''néo''(="new") and λόγος /''lógos'' meaning "speech, utterance"] is a relatively recent or isolated term, word, or phrase that may be in the process of entering common use, but that has not been fully accepted int ...
meaning "on the same ribbon"; Ancient Greek, Greek: ', ''syn'' "along with" + ', ''tainiā'' "band". This can be interpreted classically as "on the same chromosome", or in the modern sense of having the same order of genes on two (homologous) strings of DNA (or chromosomes).
: co-localization on a chromosome
The classical concept is related togenetic linkage
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separ ...
: Linkage between two loci is established by the observation of lower-than-expected recombination frequencies between them. In contrast, any loci on the same chromosome are by definition syntenic, even if their recombination frequency cannot be distinguished from unlinked loci by practical experiments. Thus, in theory, all linked loci are syntenic, but not all syntenic loci are necessarily linked. Similarly, in genomics
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dim ...
, the genetic loci on a chromosome are syntenic regardless of whether this relationship can be established by experimental methods such as DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ...
/assembly, genome walking, physical localization or hap-mapping.
Students of (classical) genetics employ the term synteny to describe the situation in which two genetic loci have been assigned to the same chromosome but still may be separated by a large enough distance in map units that genetic linkage has not been demonstrated.
Across evolution:
Shared synteny (also known as conserved synteny) describes preserved co-localization of genes on chromosomes of different species. Duringevolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation ...
, rearrangements to the genome such as chromosome translocations may separate two loci, resulting in the loss of synteny between them. Conversely, translocations can also join two previously separate pieces of chromosomes together, resulting in a gain of synteny between loci. Stronger-than-expected shared synteny can reflect selection for functional relationships between syntenic genes, such as combinations of alleles that are advantageous when inherited together, or shared regulatory mechanisms.
In light of the more recent shift in the meaning of ''synteny'', this conservation of gene content and linkage without preservation of order has also been termed ''mesosynteny''.
Current concept: gene-order preservation
 The term is currently (since ~2000) more commonly used to describe preservation of the precise order of genes on a chromosome passed down from a common ancestor, despite more "old school" geneticists rejecting what they perceive as a misappopriation of the term, preferring ''collinearity'' instead.
The analysis of synteny in the gene order sense has several applications in genomics. Shared synteny is one of the most reliable criteria for establishing the orthology of genomic regions in different species. Additionally, exceptional conservation of synteny can reflect important functional relationships between genes. For example, the order of genes in the "
The term is currently (since ~2000) more commonly used to describe preservation of the precise order of genes on a chromosome passed down from a common ancestor, despite more "old school" geneticists rejecting what they perceive as a misappopriation of the term, preferring ''collinearity'' instead.
The analysis of synteny in the gene order sense has several applications in genomics. Shared synteny is one of the most reliable criteria for establishing the orthology of genomic regions in different species. Additionally, exceptional conservation of synteny can reflect important functional relationships between genes. For example, the order of genes in the "Hox cluster
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the c ...
", which are key determinants of the animal
Animals are multicellular, eukaryotic organisms in the Kingdom (biology), biological kingdom Animalia. With few exceptions, animals Heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, are Motilit ...
body plan and which interact with each other in critical ways, is essentially preserved throughout the animal kingdom.
Synteny is widely used in studying complex genomes, as comparative genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural lan ...
allows the presence and possibly function of genes in a simpler, model organism to infer those in a more complex one. For example, wheat has a very large, complex genome which is difficult to study. In 1994 research from the John Innes Centre in England and the National Institute of Agrobiological Research in Japan demonstrated that the much smaller rice genome had a similar structure and gene order to that of wheat. Further study found that many cereals are syntenic and thus plants such as rice
Rice is the seed of the grass species ''Oryza sativa'' (Asian rice) or less commonly ''Oryza glaberrima
''Oryza glaberrima'', commonly known as African rice, is one of the two domesticated rice species. It was first domesticated and grown i ...
or the grass ''Brachypodium
''Brachypodium'' is a genus of plants in the grass family, widespread across much of Africa, Eurasia, and Latin America. The genus is classified in its own tribe Brachypodieae.
Flimsy upright stems form tussocks. Flowers appear in compact spi ...
'' could be used as a model to find genes or genetic markers of interest which could be used in wheat breeding and research. In this context, synteny was also essential in identifying a highly important region in wheat, the Ph1 locus involved in genome stability and fertility, which was located using information from syntenic regions in rice and Brachypodium.
Synteny is also widely used in microbial genomics. In Hyphomicrobiales and Enterobacteriales, syntenic genes encode a large number of essential cell functions and represent a high level of functional relationships.
Patterns of shared synteny or synteny breaks can also be used as characters to infer the phylogenetic
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups o ...
relationships among several species, and even to infer the genome organization of extinct ancestral species. A qualitative distinction is sometimes drawn between macrosynteny, preservation of synteny in large portions of a chromosome, and microsynteny, preservation of synteny for only a few genes at a time.
Computational detection
Shared synteny between different species can be inferred from their genomic sequences. This is typically done using a version of the MCScan algorithm, which finds syntenic blocks between species by comparing their homologous genes and looking for common patterns of collinearity on a chromosomal or contig scale. Homologies are usually determined on the basis of high bit score BLAST hits that occur between multiple genomes. From here,dynamic programming
Dynamic programming is both a mathematical optimization method and a computer programming method. The method was developed by Richard Bellman in the 1950s and has found applications in numerous fields, from aerospace engineering to economics.
I ...
is used to select the best scoring path of shared homologous genes between species, taking into account potential gene loss and gain which may have occurred in the species' evolutionary histories.
See also
* Ridge (biology) * Ultra-conserved element *Comparative genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural lan ...
References
{{reflistExternal links
ACT (Artemis Comparison Tool)
— Probably the most used synteny software program used in comparative genomics.
Comparative Maps
NIH's National Library of Medicine NCBI link to Gene Homology resources, and Comparative Chromosome Maps of the Human, Mouse, and Rat.
More information on synteny and its use in comparative cereal genomics.
NCBI Home Page
NIH's National Library of Medicine NCBI (National Center for Biotechnology Information) link to a tremendous number of resources.
SimpleSynteny
A free browser-based tool to compare and visualize microsynteny across multiple genomes for a set of genes.
Synteny server
Server for Synteny Identification and Analysis of Genome Rearrangement—the Identification of synteny and calculating reversal distances.
PlantSyntenyViewer
A web based visualisation tool allowing to navigate on genomes and visualizing the Synteny conservation among several datasets (cereals, dicotyledons, animals, a Wheat-based one...) Bioinformatics Classical genetics