Protein-fragment complementation assay on:
[Wikipedia]
[Google]
[Amazon]
Within the field of
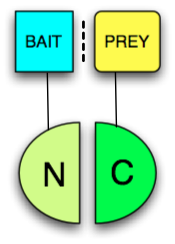 Any protein that can be split into two parts and reconstituted non-covalently to form a functional protein may be used in a PCA. The two fragments however have low affinity for each other and must be brought together by other interacting proteins fused to them (often called "bait" and "prey" since the bait protein can be used to identify a prey protein, see figure). The protein that produces a detectable readout is called "reporter". Usually enzymes which confer resistance to nutrient deprivation or antibiotics, such as dihydrofolate reductase or
Any protein that can be split into two parts and reconstituted non-covalently to form a functional protein may be used in a PCA. The two fragments however have low affinity for each other and must be brought together by other interacting proteins fused to them (often called "bait" and "prey" since the bait protein can be used to identify a prey protein, see figure). The protein that produces a detectable readout is called "reporter". Usually enzymes which confer resistance to nutrient deprivation or antibiotics, such as dihydrofolate reductase or
molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
, a protein-fragment complementation assay, or PCA, is a method for the identification and quantification of protein–protein interactions. In the PCA, the proteins of interest ("bait" and "prey") are each covalently linked to fragments of a third protein (e.g. DHFR, which acts as a "reporter"). Interaction between the bait and the prey proteins brings the fragments of the reporter protein in close proximity to allow them to form a functional reporter protein whose activity can be measured. This principle can be applied to many different reporter proteins and is also the basis for the yeast two-hybrid system, an archetypical PCA assay.
Split protein assays
 Any protein that can be split into two parts and reconstituted non-covalently to form a functional protein may be used in a PCA. The two fragments however have low affinity for each other and must be brought together by other interacting proteins fused to them (often called "bait" and "prey" since the bait protein can be used to identify a prey protein, see figure). The protein that produces a detectable readout is called "reporter". Usually enzymes which confer resistance to nutrient deprivation or antibiotics, such as dihydrofolate reductase or
Any protein that can be split into two parts and reconstituted non-covalently to form a functional protein may be used in a PCA. The two fragments however have low affinity for each other and must be brought together by other interacting proteins fused to them (often called "bait" and "prey" since the bait protein can be used to identify a prey protein, see figure). The protein that produces a detectable readout is called "reporter". Usually enzymes which confer resistance to nutrient deprivation or antibiotics, such as dihydrofolate reductase or beta-lactamase
Beta-lactamases, (β-lactamases) are enzymes () produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems ( ertapenem), although carbap ...
respectively, or proteins that give colorimetric or fluorescent signals are used as reporters. When fluorescent proteins are reconstituted the PCA is called Bimolecular fluorescence complementation assay. The following proteins have been used in split protein PCAs:
*Beta-lactamase
Beta-lactamases, (β-lactamases) are enzymes () produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems ( ertapenem), although carbap ...
* Dihydrofolate reductase (DHFR)
*Focal adhesion kinase
PTK2 protein tyrosine kinase 2 (PTK2), also known as focal adhesion kinase (FAK), is a protein that, in humans, is encoded by the ''PTK2'' gene. PTK2 is a focal adhesion-associated protein kinase involved in cellular adhesion (how cells stick to ...
(FAK)
*Gal4, a yeast transcription factor (as in the classical yeast two-hybrid system)
*GFP (split-GFP), e.g. EGFP (enhanced green fluorescent protein)
* Horseradish peroxidase
*Infrared fluorescent protein IFP1.4, an engineered chromophore-binding domain (CBD) of a bacteriophytochrome from ''Deinococcus radiodurans
''Deinococcus radiodurans'' is an extremophilic bacterium and one of the most radiation-resistant organisms known. It can survive cold, dehydration, vacuum, and acid, and therefore is known as a polyextremophile. It has been listed as the world' ...
''
*LacZ (beta-galactosidase
β-Galactosidase (EC 3.2.1.23, lactase, beta-gal or β-gal; systematic name β-D-galactoside galactohydrolase), is a glycoside hydrolase enzyme that catalyzes hydrolysis of terminal non-reducing β-D-galactose residues in β-D-galactosides.
β- ...
)
* Luciferase, including ReBiL (recombinase enhanced bimolecular luciferase) and '' Gaussia princeps'' luciferase. Commercial products using luciferase include NanoLuc and NanoBIT. A modification has also been developed for lipid droplet-associated interactions.
*TEV (Tobacco etch virus
Tobacco etch virus (TEV) is a plant virus in the genus ''Potyvirus'' and family ''Potyviridae''. Like other members of the genus ''Potyvirus'', TEV has a monopartite positive-sense, single-stranded RNA genome surrounded by a capsid made from a ...
protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the ...
)
*Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
Genome-wide applications
The methods mentioned above have been applied to wholegenome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding g ...
s, e.g. yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constit ...
or syphilis bacteria.
References
Further reading
* {{refend Molecular biology Laboratory techniques