Methyl transferase on:
[Wikipedia]
[Google]
[Amazon]

 Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a
RCC1
an important
 Histone methyltransferases are critical for genetic regulation at the epigenetic level. They modify mainly lysine on the ε-nitrogen and the arginine
Histone methyltransferases are critical for genetic regulation at the epigenetic level. They modify mainly lysine on the ε-nitrogen and the arginine
 N-alpha methyltransferases transfer a methyl group from SAM to the N-terminal nitrogen on protein targets. The N-terminal methionine is first cleaved by another enzyme and the X- Proline-Lysine consensus sequence is recognized by the methyltransferase. For all known substrates, the X amino acid is
N-alpha methyltransferases transfer a methyl group from SAM to the N-terminal nitrogen on protein targets. The N-terminal methionine is first cleaved by another enzyme and the X- Proline-Lysine consensus sequence is recognized by the methyltransferase. For all known substrates, the X amino acid is
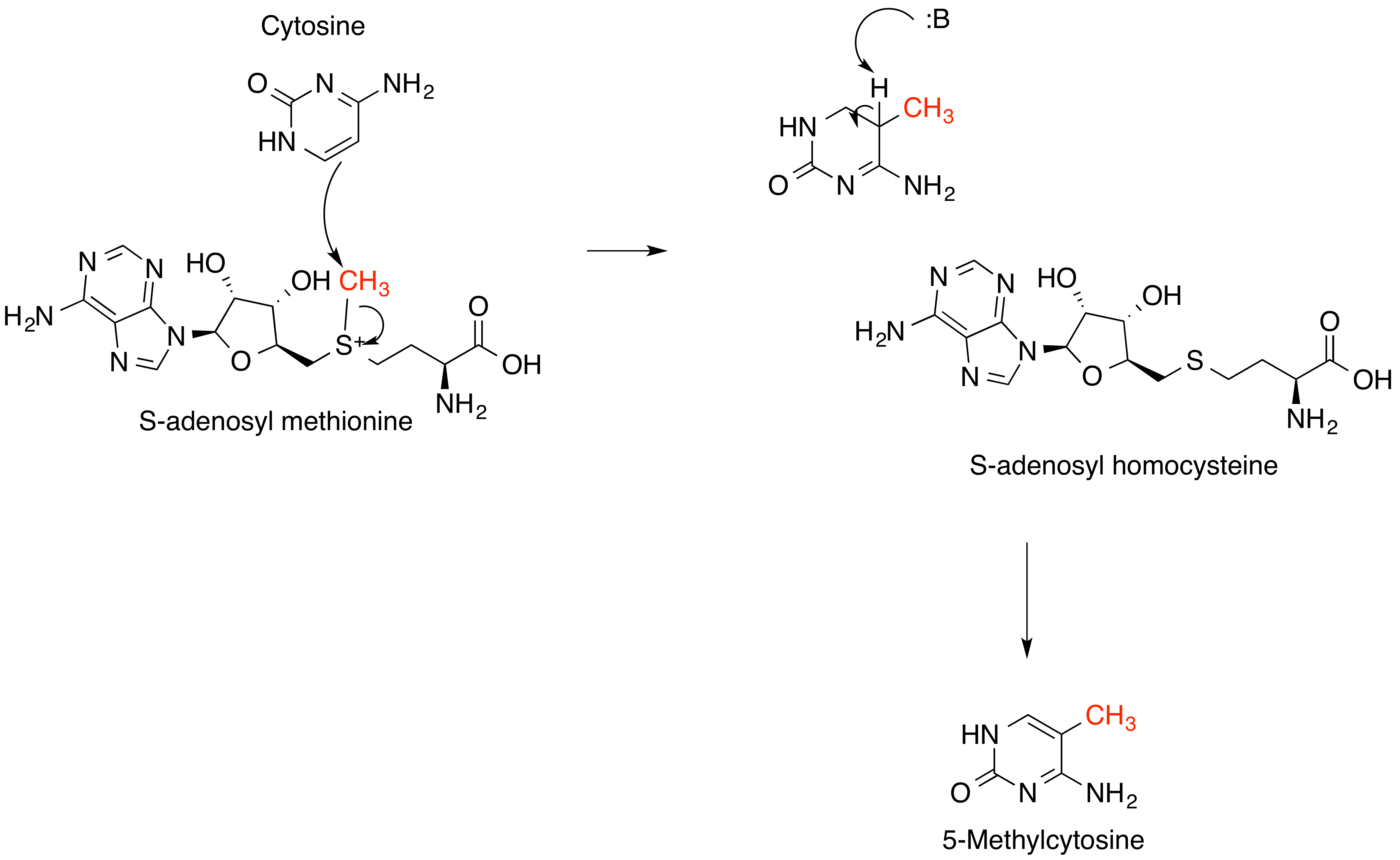
 DNA methylation, a key component of genetic regulation, occurs primarily at the 5-carbon of the base
DNA methylation, a key component of genetic regulation, occurs primarily at the 5-carbon of the base
 Natural product methyltransferases (NPMTs) are a diverse group of enzymes that add methyl groups to naturally-produced small molecules. Like many methyltransferases, SAM is utilized as a methyl donor and SAH is produced. Methyl groups are added to S, N, O, or C atoms, and are classified by which of these atoms are modified, with O-methyltransferases representing the largest class. The methylated products of these reactions serve a variety of functions, including co-factors, pigments, signalling compounds, and metabolites. NPMTs can serve a regulatory role by modifying the reactivity and availability of these compounds. These enzymes are not highly conserved across different species, as they serve a more specific function in providing small molecules for specialized pathways in species or smaller groups of species. Reflective of this diversity is the variety of catalytic strategies, including general acid-base catalysis
Natural product methyltransferases (NPMTs) are a diverse group of enzymes that add methyl groups to naturally-produced small molecules. Like many methyltransferases, SAM is utilized as a methyl donor and SAH is produced. Methyl groups are added to S, N, O, or C atoms, and are classified by which of these atoms are modified, with O-methyltransferases representing the largest class. The methylated products of these reactions serve a variety of functions, including co-factors, pigments, signalling compounds, and metabolites. NPMTs can serve a regulatory role by modifying the reactivity and availability of these compounds. These enzymes are not highly conserved across different species, as they serve a more specific function in providing small molecules for specialized pathways in species or smaller groups of species. Reflective of this diversity is the variety of catalytic strategies, including general acid-base catalysis
metal-based catalysis
and proximity and desolvation effects not requiring catalytic amino acids. NPMTs are the most functionally diverse class of methyltransferases. Important examples of this enzyme class in humans include
Important examples of this enzyme class in humans include

3-D Structure of DNA Methyltransferase
A novel methyltransferase : the 7SK snRNA Methylphosphate Capping Enzyme
as seen on Flintbox
"The Role of Methylation in Gene Expression"
on Nature Scitable
"Nutrition and Depression: Nutrition, Methylation, and Depression"
on Psychology Today
"DNA Methylation - What is DNA Methylation?"
from News-Medical.net
Genetic pathways involving Histone Methyltransferases from Cell Signaling Technology {{Portal bar, Biology, border=no EC 2.1.1 Methylation

 Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold
The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonde ...
for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile
In chemistry, an electrophile is a chemical species that forms bonds with nucleophiles by accepting an electron pair. Because electrophiles accept electrons, they are Lewis acids. Most electrophiles are positively charged, have an atom that carrie ...
that transfers the methyl group to the enzyme substrate. SAM is converted to ''S''-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
Function
Genetics
Methylation, as well as other epigenetic modifications, affectstranscription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
, gene stability, and parental imprinting. It directly impacts chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in r ...
structure and can modulate gene transcription, or even completely silence
Silence is the absence of ambient audible sound, the emission of sounds of such low intensity that they do not draw attention to themselves, or the state of having ceased to produce sounds; this latter sense can be extended to apply to the c ...
or activate genes, without mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
to the gene itself. Though the mechanisms of this genetic control are complex, hypo- and hypermethylation of DNA is implicated in many diseases.
Protein regulation
Methylation of proteins has a regulatory role inprotein–protein interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and th ...
s, protein–DNA interaction
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, becaus ...
s, and protein activation.
ExamplesRCC1
an important
mitotic
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintai ...
protein, is methylated so that it can interact with centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers ...
s of chromosomes. This is an example of regulation of protein-protein interaction, as methylation regulates the attachment of RCC1 to histone proteins H2A and H2B. The RCC1-chromatin interaction is also an example of a protein-DNA interaction, as another domain of RCC1 interacts directly with DNA when this protein is methylated. When RCC1 is not methylated, dividing cells have multiple spindle poles and usually cannot survive.
p53
p53, also known as Tumor protein P53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory protein that is often mutated in human cancers. The p53 proteins (originally thought to be, and often s ...
methylated on lysine to regulate its activation and interaction with other proteins in the DNA damage response. This is an example of regulation of protein-protein interactions and protein activation. p53 is a known tumor suppressor
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or re ...
that activates DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA da ...
pathways, initiates apoptosis, and pauses the cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and sub ...
. Overall, it responds to mutations in DNA, signaling to the cell to fix them or to initiate cell death so that these mutations cannot contribute to cancer.
NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a protein complex that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular ...
(a protein involved in inflammation) is a known methylation target of the methyltransferase SETD6
SET domain containing 6 is a protein in humans that is encoded by the SETD6 gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''b ...
, which turns off NF-κB signaling by inhibiting of one of its subunits, RelA
Transcription factor p65 also known as nuclear factor NF-kappa-B p65 subunit is a protein that in humans is encoded by the ''RELA'' gene.
RELA, also known as p65, is a REL-associated protein involved in NF-κB heterodimer formation, nuclear tra ...
. This reduces the transcriptional activation and inflammatory response
Inflammation (from la, inflammatio) is part of the complex biological response of body tissues to harmful stimuli, such as pathogens, damaged cells, or irritants, and is a protective response involving immune cells, blood vessels, and molecu ...
, making methylation of NF-κB a regulatory process by which cell signaling through this pathway is reduced.
Natural product methyltransferases provide a variety of inputs into metabolic pathways, including the availability of cofactors, signalling molecules, and metabolites. This regulates various cellular pathways by controlling protein activity.
Types
Histone methyltransferases
 Histone methyltransferases are critical for genetic regulation at the epigenetic level. They modify mainly lysine on the ε-nitrogen and the arginine
Histone methyltransferases are critical for genetic regulation at the epigenetic level. They modify mainly lysine on the ε-nitrogen and the arginine guanidinium
Guanidine is the compound with the formula HNC(NH2)2. It is a colourless solid that dissolves in polar solvents. It is a strong base that is used in the production of plastics and explosives. It is found in urine predominantly in patients experie ...
group on histone tails. Lysine methyltransferases and Arginine methyltransferases are unique classes of enzymes, but both bind SAM as a methyl donor for their histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
substrates. Lysine amino acids can be modified with one, two, or three methyl groups, while Arginine amino acids can be modified with one or two methyl groups. This increases the strength of the positive charge and residue hydrophobicity
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water.
Hydrophobic molecules tend to be nonpolar and, t ...
, allowing other proteins to recognize methyl marks. The effect of this modification depends on the location of the modification on the histone tail and the other histone modifications around it. The location of the modifications can be partially determined by DNA sequence, as well as small non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally im ...
s and the methylation of the DNA itself. Most commonly, it is histone H3 or H4 that is methylated in vertebrates. Either increased or decreased transcription of genes around the modification can occur. Increased transcription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
is a result of decreased chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in r ...
condensation, while decreased transcription results from increased chromatin condensation. Methyl marks on the histones contribute to these changes by serving as sites for recruitment of other proteins that can further modify chromatin.
N-terminal methyltransferases
 N-alpha methyltransferases transfer a methyl group from SAM to the N-terminal nitrogen on protein targets. The N-terminal methionine is first cleaved by another enzyme and the X- Proline-Lysine consensus sequence is recognized by the methyltransferase. For all known substrates, the X amino acid is
N-alpha methyltransferases transfer a methyl group from SAM to the N-terminal nitrogen on protein targets. The N-terminal methionine is first cleaved by another enzyme and the X- Proline-Lysine consensus sequence is recognized by the methyltransferase. For all known substrates, the X amino acid is Alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side ...
, Serine, or Proline. This reaction yields a methylated protein and SAH. Known targets of these methyltransferases in humans include RCC-1 (a regulator of nuclear transport proteins) and Retinoblastoma protein
The retinoblastoma protein (protein name abbreviated pRb; gene name abbreviated ''Rb'', ''RB'' or ''RB1'') is a proto-oncogenic tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive ...
(a tumor suppressor protein that inhibits excessive cell division). RCC-1 methylation is especially important in mitosis as it coordinates the localization of some nuclear
Nuclear may refer to:
Physics
Relating to the nucleus of the atom:
* Nuclear engineering
*Nuclear physics
*Nuclear power
*Nuclear reactor
*Nuclear weapon
*Nuclear medicine
*Radiation therapy
*Nuclear warfare
Mathematics
*Nuclear space
*Nuclear ...
proteins in the absence of the nuclear envelope. When RCC-1 is not methylated, cell division is abnormal following the formation of extra spindle poles. The function of Retinoblastoma protein N-terminal methylation is not known.
DNA/RNA methyltransferases
 DNA methylation, a key component of genetic regulation, occurs primarily at the 5-carbon of the base
DNA methylation, a key component of genetic regulation, occurs primarily at the 5-carbon of the base cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an ...
, forming 5’methylcytosine (see left). Methylation is an epigenetic modification catalyzed by DNA methyltransferase enzymes, including DNMT1, DNMT2 (renamed TRDMT1 to reflect its function methylating tRNA, not DNA), and DNMT3. These enzymes use S-adenosylmethionine as a methyl donor and contain several highly conserved structural features between the three forms; these include the S-adenosylmethionine binding site, a vicinal proline-cysteine pair which forms a thiolate anion important for the reaction mechanism, and the cytosine substrate binding pocket. Many features of DNA methyltransferases are highly conserved throughout many classes of life, from bacteria to mammals. In addition to controlling the expression of certain genes, there are a variety of protein complexes, many with implications for human health, which only bind to methylated DNA recognition sites. Many of the early DNA methyltransferases have been thought to be derived from RNA methyltransferases that were supposed to be active in the RNA world
The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existen ...
to protect many species of primitive RNA.
RNA methylation has been observed in different types of RNA species viz.mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
, rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosoma ...
, tRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
, snoRNA
In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, ...
, snRNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcri ...
, miRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miR ...
, tmRNA
Transfer-messenger RNA (abbreviated tmRNA, also known as 10Sa RNA and by its genetic name SsrA) is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with ...
as well as viral RNA species. Specific RNA methyltransferases are employed by cells to mark these on the RNA species according to the need and environment prevailing around the cells, which form a part of field called molecular epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are ...
. 2'-O-methylation, m6A methylation, m1G methylation as well as m5C are most commonly methylation marks observed in different types of RNA.
6A is an enzyme that catalyzes chemical reaction as following:
S-adenosyl-L-methionine + DNA adenine S-adenosyl-L-homocysteine + DNA 6-methylaminopurine
m6A was primarily found in prokaryotes until 2015 when it was also identified in some eukaryotes. m6A methyltransferases methylate the amino group in DNA at C-6 position specifically to prevent the host system to digest own genome through restriction enzymes.
m5C plays a role to regulate gene transcription. m5C transferases are the enzymes that produce C5-methylcytosine in DNA at C-5 position of cytosine and are found in most plants and some eukaryotes.
Natural product methyltransferases
 Natural product methyltransferases (NPMTs) are a diverse group of enzymes that add methyl groups to naturally-produced small molecules. Like many methyltransferases, SAM is utilized as a methyl donor and SAH is produced. Methyl groups are added to S, N, O, or C atoms, and are classified by which of these atoms are modified, with O-methyltransferases representing the largest class. The methylated products of these reactions serve a variety of functions, including co-factors, pigments, signalling compounds, and metabolites. NPMTs can serve a regulatory role by modifying the reactivity and availability of these compounds. These enzymes are not highly conserved across different species, as they serve a more specific function in providing small molecules for specialized pathways in species or smaller groups of species. Reflective of this diversity is the variety of catalytic strategies, including general acid-base catalysis
Natural product methyltransferases (NPMTs) are a diverse group of enzymes that add methyl groups to naturally-produced small molecules. Like many methyltransferases, SAM is utilized as a methyl donor and SAH is produced. Methyl groups are added to S, N, O, or C atoms, and are classified by which of these atoms are modified, with O-methyltransferases representing the largest class. The methylated products of these reactions serve a variety of functions, including co-factors, pigments, signalling compounds, and metabolites. NPMTs can serve a regulatory role by modifying the reactivity and availability of these compounds. These enzymes are not highly conserved across different species, as they serve a more specific function in providing small molecules for specialized pathways in species or smaller groups of species. Reflective of this diversity is the variety of catalytic strategies, including general acid-base catalysismetal-based catalysis
and proximity and desolvation effects not requiring catalytic amino acids. NPMTs are the most functionally diverse class of methyltransferases.
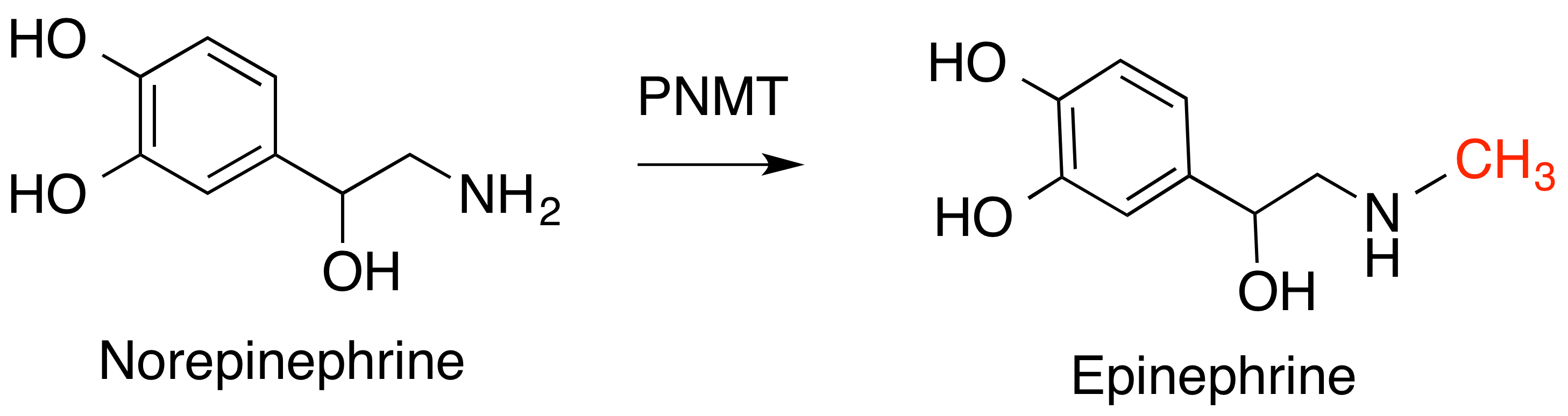
phenylethanolamine N-methyltransferase
Phenylethanolamine ''N''-methyltransferase (PNMT) is an enzyme found primarily in the adrenal medulla that converts norepinephrine (noradrenaline) to epinephrine (adrenaline). It is also expressed in small groups of neurons in the human brain and ...
(PNMT), which converts norepinephrine
Norepinephrine (NE), also called noradrenaline (NA) or noradrenalin, is an organic chemical in the catecholamine family that functions in the brain and body as both a hormone and neurotransmitter. The name "noradrenaline" (from Latin '' ad' ...
to epinephrine, and histamine N-methyltransferase
Histamine ''N''-methyltransferase (HNMT, HMT) is an enzyme involved in the metabolism of histamine. It is one of two enzymes involved in the metabolism of histamine in mammals, the other being diamine oxidase (DAO). HNMT catalyzes the methylation ...
(HNMT), which methylates histamine
Histamine is an organic nitrogenous compound involved in local immune responses, as well as regulating physiological functions in the gut and acting as a neurotransmitter for the brain, spinal cord, and uterus. Since histamine was discovered ...
in the process of histamine metabolism. Catechol-''O''-methyltransferase (COMT) degrades a class of molecules known as catcholamines that includes dopamine, epinephrine, and norepenepherine.
Non-SAM dependent methyltransferases
Methanol, methyl tetrahydrofolate,mono-
Numeral or number prefixes are prefixes derived from numerals or occasionally other numbers. In English and many other languages, they are used to coin numerous series of words. For example:
* unicycle, bicycle, tricycle (1-cycle, 2-cycle, 3-cy ...
, di-, and trimethylamine
Trimethylamine (TMA) is an organic compound with the formula N(CH3)3. It is a colorless, hygroscopic, and flammable tertiary amine. It is a gas at room temperature but is usually sold as a 40% solution in water. (It is also sold in pressurized ...
, methanethiol
Methanethiol (also known as methyl mercaptan) is an organosulfur compound with the chemical formula . It is a colorless gas with a distinctive putrid smell. It is a natural substance found in the blood, brain and feces of animals (including humans ...
, methyltetrahydromethanopterin, and chloromethane
Chloromethane, also called methyl chloride, Refrigerant-40, R-40 or HCC 40, is an organic compound with the chemical formula . One of the haloalkanes, it is a colorless, odorless, flammable gas. Methyl chloride is a crucial reagent in industrial ...
are all methyl donors found in biology as methyl group donors, typically in enzymatic reactions using the cofactor vitamin B12
Vitamin B12, also known as cobalamin, is a water-soluble vitamin involved in metabolism. It is one of eight B vitamins. It is required by animals, which use it as a cofactor in DNA synthesis, in both fatty acid and amino acid metabolism. ...
. These substrates contribute to methyl transfer pathways including methionine biosynthesis, methanogenesis, and acetogenesis Acetogenesis is a process through which acetate is produced either by the reduction of CO2 or by the reduction of organic acids, rather than by the oxidative breakdown of carbohydrates or ethanol, as with acetic acid bacteria.
The different bac ...
.
Radical SAM methyltransferases
Based on different protein structures and mechanisms of catalysis, there are 3 different types of radical SAM (RS) methylases: Class A, B, and C. Class A RS methylases are the best characterized of the 4 enzymes and are related to both RlmN and Cfr. RlmN is ubiquitous in bacteria which enhances translational fidelity and RlmN catalyzes methylation of C2 of adenosine 2503 (A2503) in 23 S rRNA and C2 of adenosine (A37). Cfr, on the other hand, catalyzes methylation of C8 of A2503 as well and it also catalyzes C2 methylation. Class B is currently the largest class of radical SAM methylases which can methylate both ''sp''2-hybridized and ''sp''3-hybridized carbon atoms in different sets of substrates unlike Class A which only catalyzes ''sp''2-hybridized carbon atoms. The main difference that distinguishes Class B from others is the additional N-terminal cobalamin-binding domain that binds to the RS domain. Class C methylase has homologous sequence with the RS enzyme, coproporphyrinogen III oxidase (HemN), which also catalyzes the methylation of ''sp''2-hybridized carbon centers yet it lacks the 2 cysteines required for methylation in mechanism of Class A.
Clinical significance
As with any biological process which regulates gene expression and/or function, anomalous DNA methylation is associated with genetic disorders such as ICF,Rett syndrome
Rett syndrome (RTT) is a genetic disorder that typically becomes apparent after 6–18 months of age and almost exclusively in females. Symptoms include impairments in language and coordination, and repetitive movements. Those affected often h ...
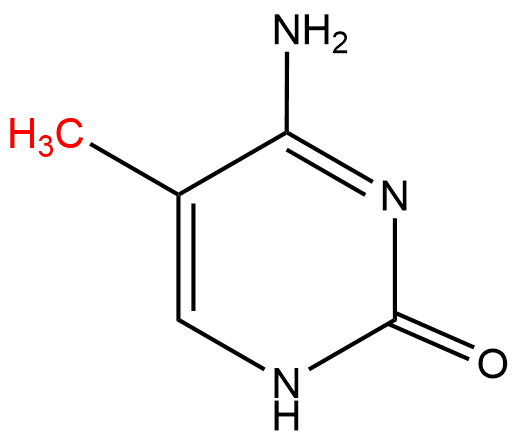
, and Fragile X syndrome. Cancer cells typically exhibit less DNA methylation activity in general, though often hypermethylation at sites which are unmethylated in normal cells; this overmethylation often functions as a way to inactivate tumor-suppressor genes. Inhibition of overall DNA methyltransferase activity has been proposed as a treatment option, but DNMT inhibitors, analogs of their cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an ...
substrates, have been found to be highly toxic due to their similarity to cytosine (see right); this similarity to the nucleotide causes the inhibitor to be incorporated into DNA translation, causing non-functioning DNA to be synthesized.
A methylase which alters the ribosomal RNA binding site of the antibiotic linezolid
Linezolid is an antibiotic used for the treatment of infections caused by Gram-positive bacteria that are resistant to other antibiotics. Linezolid is active against most Gram-positive bacteria that cause disease, including streptococci, v ...
causes cross-resistance to other antibiotics that act on the ribosomal RNA. Plasmid vectors capable of transmitting this gene are a cause of potentially dangerous cross resistance.
Examples of methyltransferase enzymes relevant to disease:
*thiopurine methyltransferase
Thiopurine methyltransferase or thiopurine S-methyltransferase (TPMT) is an enzyme that in humans is encoded by the ''TPMT'' gene. A pseudogene for this locus is located on chromosome 18q.
Function
Thiopurine methyltransferase methylates t ...
: defects in this gene causes toxic accumulation of thiopurine compounds, drugs used in chemotherapy and immunosuppressant therapy
*methionine synthase
Methionine synthase also known as MS, MeSe, MTR is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the ''MTR'' gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms ...
: pernicious anemia
Pernicious anemia is a type of vitamin B12 deficiency anemia, a disease in which not enough red blood cells are produced due to the malabsorption of vitamin B12. Malabsorption in pernicious anemia results from the lack or loss of intrinsic fa ...
, caused by Vitamin B12
Vitamin B12, also known as cobalamin, is a water-soluble vitamin involved in metabolism. It is one of eight B vitamins. It is required by animals, which use it as a cofactor in DNA synthesis, in both fatty acid and amino acid metabolism. ...
deficiency, is caused by a lack of cofactor for the methionine synthase enzyme
Applications in drug discovery and development
Recent work has revealed the methyltransferases involved in methylation of naturally occurring anticancer agents to useS-Adenosyl methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throug ...
(SAM) analogs that carry alternative alkyl groups as a replacement for methyl. The development of the facile chemoenzymatic platform to generate and utilize differentially alkylated SAM analogs in the context of drug discovery and drug development
Drug development is the process of bringing a new pharmaceutical drug to the market once a lead compound has been identified through the process of drug discovery. It includes preclinical research on microorganisms and animals, filing for re ...
is known as alkylrandomization.
Applications in cancer treatment
In human cells, it was found that m5C was associated with abnormal tumor cells in cancer. The role and potential application of m5C includes to balance the impaired DNA in cancer both hypermethylation and hypomethylation. An epigenetic repair of DNA can be applied by changing the m5C amount in both types of cancer cells (hypermethylation/ hypomethylation) and as well as the environment of the cancers to reach an equivalent point to inhibit tumor cells.Examples
Examples include: *Catechol-O-methyltransferase
Catechol-''O''-methyltransferase (COMT; ) is one of several enzymes that degrade catecholamines (neurotransmitters such as dopamine, epinephrine, and norepinephrine), catecholestrogens, and various drugs and substances having a catechol struct ...
* DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl m ...
* Histone methyltransferase
* 5-Methyltetrahydrofolate-homocysteine methyltransferase
Methionine synthase also known as MS, MeSe, MTR is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the ''MTR'' gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms ...
* O-methyltransferase
* methionine synthase
Methionine synthase also known as MS, MeSe, MTR is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the ''MTR'' gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms ...
* corrinoid-iron sulfur protein
References
Further reading
*3-D Structure of DNA Methyltransferase
A novel methyltransferase : the 7SK snRNA Methylphosphate Capping Enzyme
as seen on Flintbox
"The Role of Methylation in Gene Expression"
on Nature Scitable
"Nutrition and Depression: Nutrition, Methylation, and Depression"
on Psychology Today
"DNA Methylation - What is DNA Methylation?"
from News-Medical.net
Genetic pathways involving Histone Methyltransferases from Cell Signaling Technology {{Portal bar, Biology, border=no EC 2.1.1 Methylation