LIG1 on:
[Wikipedia]
[Google]
[Amazon]
DNA ligase 1 is an

 During
During
 DNA ligase I functions to ligate single stranded DNA breaks in the final step of the
DNA ligase I functions to ligate single stranded DNA breaks in the final step of the
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
that in humans is encoded by the ''LIG1'' gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
. DNA ligase I is the only known eukaryotic DNA ligase involved in both DNA replication and repair, making it the most studied of the ligase
In biochemistry, a ligase is an enzyme that can catalyze the joining (ligation) of two large molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the larger molecules or the enzym ...
s.
Discovery
It was known that DNA replication occurred through the breakage of the double DNA strand, but the enzyme responsible for ligating the strands back together, and mechanism of action, was unknown until Lehman, Gellert, Richardson, and Hurwitz laboratories, made significant contributions to the discovery of DNA ligase in 1967.
Recruitment and regulation
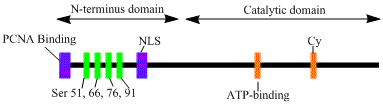
The LIG1 gene encodes a, 120kDa enzyme, 919 residues long, known as DNA ligase I. The DNA ligase I polypeptide contains anN-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
replication factory-targeting sequence (RFTS), followed by a nuclear localization sequence A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines ...
(NLS), and three functional domains. The three domains consist of an N-terminal DNA binding domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a genera ...
(DBD), and catalytic nucleotidyltransferase
Nucleotidyltransferases are transferase enzymes of phosphorus-containing groups, e.g., substituents of nucleotidylic acids or simply nucleoside monophosphates. The general reaction of transferring a nucleoside monophosphate moiety from A to B, can ...
(NTase), and C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
/ oligosaccharide
An oligosaccharide (/ˌɑlɪgoʊˈsækəˌɹaɪd/; from the Greek ὀλίγος ''olígos'', "a few", and σάκχαρ ''sácchar'', "sugar") is a saccharide polymer containing a small number (typically two to ten) of monosaccharides (simple sug ...
binding (OB) domains. Although the N-terminus of the peptide has no catalytic activity it is needed for activity within the cells. The N-terminus of the protein contains a replication factory-targeting sequence that is used to recruit it to sites of DNA replication known as replication factories.
Activation and recruitment of DNA ligase I seem to be associated with posttranslational modifications. N-terminal domain is completed through phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
of four serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − for ...
residues on this domain, Ser51, Ser76, and Ser91 by cyclin-dependent kinase
Cyclin-dependent kinases (CDKs) are the families of protein kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. They ...
(CDK) and Ser66 by casein kinase II (CKII). Phosphorylation of these residues (Ser66 in particular) has been shown to possibly regulate the interaction between the RFTS to the proliferating cell nuclear antigen
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, wher ...
(PCNA) when ligase I is recruited to the replication factories during S-phase
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during ...
. Rossi et al. proposed that when Ser66 is dephosphorylated, the RFTS of ligase I interact with PCNA, which was confirmed in vitro by Tom et al. Both data sets provide plausible evidence the N-terminal region of ligase I plays a regulatory role in the enzymes in vivo function in the nucleus. Moreover, the identification of a cyclin binding (Cy) motif in the catalytic C-terminus domain was shown by mutational analysis to play a role in the phosphorylation of serines 91 and 76. Together, the N-terminal serines are substrates of the CDK and CKII, which appear to play an important regulatory role DNA ligase I recruitment to the replication factory during S-phase of the cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and sub ...
.

Function and mechanism
LIG1 encodes DNA ligase I, which functions in DNA replication and thebase excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from ...
process.
Eukaryotic DNA ligase 1 catalyzes a reaction that is chemically universal to all ligases. DNA ligase 1 utilizes adenosine triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms ...
(ATP) to catalyze the energetically favorable ligation events in both DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
and repair
The technical meaning of maintenance involves functional checks, servicing, repairing or replacing of necessary devices, equipment, machinery, building infrastructure, and supporting utilities in industrial, business, and residential installa ...
. During the synthesis phase
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during ...
(S-phase) of the eukaryotic cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and sub ...
, DNA replication occurs. DNA ligase 1 is responsible for joining Okazaki fragments
Okazaki fragments are short sequences of DNA nucleotides (approximately 150 to 200 base pairs long in eukaryotes) which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA ...
formed during discontinuous DNA synthesis on the DNA's lagging strand after DNA polymerase δ has replaced the RNA primer nucleotides with DNA nucleotides. If the Okazaki fragments are not properly ligated together, the unligated DNA (containing a ‘nick’) could easily degrade to a double strand break
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dama ...
, a phenomenon known to cause genetic mutations. In order to ligate these fragments together, the ligase progresses through three steps:
# Addition of an adenosine monophosphate
Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine; it is an ester of phosphoric acid and the nucleoside adenosine. As a substit ...
(AMP) group to the enzyme, referred to as adenylylation,
# Adenosine monophosphate transfer to the DNA and
# Nick sealing, or phosphodiester bond formation.
 During
During adenylylation
Adenylylation, more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein. This covalent addition of AMP to a hydroxyl side chain of the prote ...
, there is a nucleophilic attack on the alpha phosphate of ATP from a catalytic lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated − ...
resulting in the production of inorganic pyrophosphate
In chemistry, pyrophosphates are phosphorus oxyanions that contain two phosphorus atoms in a P–O–P linkage. A number of pyrophosphate salts exist, such as disodium pyrophosphate (Na2H2P2O7) and tetrasodium pyrophosphate (Na4P2O7), among othe ...
(PPi) and a covalently bound lysine-AMP intermediate in the active site of DNA ligase 1.
During the AMP transfer step, the DNA ligase becomes associated with the DNA, locates a nick and catalyzes a reaction at the 5’ phosphate site of the DNA nick. An anionic oxygen on the 5’ phosphate of the DNA nick serves as the nucleophile, attacking the alpha phosphate of the covalently bound AMP causing the AMP to be covalently bound intermediate (DNA-AMP intermediate).
In order for the phosphodiester bond to be formed, the DNA-AMP intermediate must be cleaved off. To accomplish this task, there is a nucleophilic attack on the 5’-phosphate from the upstream 3’-hydroxyl which results in the formation of the phosphodiester bond. During this nucleophilic attack, the AMP group is pushed off the 5’ phosphate as the leaving group allowing for the nick to seal and the AMP to be released, completing one cycle of DNA ligation.
Under suboptimal conditions the ligase can disassociate from the DNA before the full reaction is complete. It has been shown that magnesium
Magnesium is a chemical element with the symbol Mg and atomic number 12. It is a shiny gray metal having a low density, low melting point and high chemical reactivity. Like the other alkaline earth metals (group 2 of the periodic ...
levels can slow the nick sealing process, causing the ligase to disassociate from the DNA, leaving an aborted adenylylated intermediate incapable of being fixed without the aid of a phosphodiesterase
A phosphodiesterase (PDE) is an enzyme that breaks a phosphodiester bond. Usually, ''phosphodiesterase'' refers to cyclic nucleotide phosphodiesterases, which have great clinical significance and are described below. However, there are many ot ...
. Aprataxin
Aprataxin is a protein that in humans is encoded by the ''APTX'' gene.
This gene encodes a member of the histidine triad (HIT) superfamily, some of which have nucleotide-binding and diadenosine polyphosphate hydrolase activities. The encoded pro ...
(a phosphodiesterase) has been shown to act on aborted DNA intermediates via hydrolysis of the AMP-phosphate bond, restoring the DNA to its initial state before the ligase had reacted.
Role in damaged base repair
base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from ...
(BER) pathway. The nitrogenous bases of DNA are commonly damaged by environmental hazards such as reactive oxygen species
In chemistry, reactive oxygen species (ROS) are highly reactive chemicals formed from diatomic oxygen (). Examples of ROS include peroxides, superoxide, hydroxyl radical, singlet oxygen, and alpha-oxygen.
The reduction of molecular oxygen () p ...
, toxins, and ionizing radiation
Ionizing radiation (or ionising radiation), including nuclear radiation, consists of subatomic particles or electromagnetic waves that have sufficient energy to ionize atoms or molecules by detaching electrons from them. Some particles can travel ...
. BER is the major repair pathway responsible for excising and replacing damaged bases. Ligase I is involved in the LP-BER pathway, whereas ligase III is involved in the major SN-BER pathway(2). LP-BER proceeds in 4 catalytic steps. First, a DNA glycosylase
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first st ...
cleaves the N-glycosidic bond, releasing the damaged base and creating an AP site– a site that lacks a purine
Purine is a heterocyclic aromatic organic compound that consists of two rings ( pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines ...
or pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The othe ...
base. In the next step, an AP endonuclease creates a nick at the 5' end of the AP site, generating a hanging deoxyribose
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of a hydroxy group. D ...
phosphate (dRP) residue in place of the AP site. DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to crea ...
then synthesizes several new bases in the 5' to 3' direction, generating a hanging stretch of DNA with the dRP at its 5' end. It is at this step that SN-BER and LP-BER diverge in mechanism – in SNBER, only a single nucleotide is added and DNA Polymerase acts as a lyase to excise the AP site. In LP-BER, several bases are synthesized, generating a hanging flap of DNA, which is cleaved by a flap endonuclease Flap endonucleases (FENs, also known as 5' durgs in older references) are a class of nucleolytic enzymes that act as both 5'-3' exonucleases and structure-specific endonucleases on specialised DNA structures that occur during the biological proces ...
. This leaves behind a nicked DNA strand that is sensed and ligated by DNA ligase. The action of ligase I is stimulated by other LP-BER enzymes, particularly AP-endonuclease and DNA polymerase.
Clinical significance
Mutations in LIG1 that lead to DNA ligase I deficiency result inimmunodeficiency
Immunodeficiency, also known as immunocompromisation, is a state in which the immune system's ability to fight infectious diseases and cancer is compromised or entirely absent. Most cases are acquired ("secondary") due to extrinsic factors that a ...
and increased sensitivity to DNA-damaging agents.
There are rare reports of patients exhibiting ligase I deficiency which resulted from inherited mutant alleles. The first case manifested as stunted growth and development and an immunodeficiency. A mouse model was made based on cell lines derived from the patient, confirming that the mutant ligase confers replication errors leading to genomic instability
Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneu ...
. Notably the mutant mice also showed increases in tumorigenesis
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abnor ...
. Molecular, cellular, and clinical features of 5 patients from 3 kindreds with biallelic mutations were reported. The patients exhibited hypogammaglobulinemia, lymphopenia, increased proportions of circulating γδT cells, and very large red cells (macrocytosis.) Clinical severity ranged from a mild antibody deficiency to a combined immunodeficiency requiring hematopoietic stem cell transplantation. Chemical and radiation defects were demonstrated to impair the DNA repair pathways. Defects in DNA ligase 1 can thus lead to different forms of autosomal recessive, partial DNA ligase 1 deficiency leading to an immunodeficiency of variable severity.
Ligase I has also been found to be upregulated in proliferating tumor cells, as opposed to benign tumor cell lines and normal human cells. Furthermore, it has been shown that inhibiting ligase I expression in these cells can have a cytotoxic effect, suggesting that ligase I inhibitors may be viable chemotherapeutic agents.
Deficiencies in aprataxin
Aprataxin is a protein that in humans is encoded by the ''APTX'' gene.
This gene encodes a member of the histidine triad (HIT) superfamily, some of which have nucleotide-binding and diadenosine polyphosphate hydrolase activities. The encoded pro ...
, a phosphodiesterase
A phosphodiesterase (PDE) is an enzyme that breaks a phosphodiester bond. Usually, ''phosphodiesterase'' refers to cyclic nucleotide phosphodiesterases, which have great clinical significance and are described below. However, there are many ot ...
responsible for reconditioning the DNA (after DNA ligase I aborts the adenylylated DNA intermediate), has been linked to neurodegeneration
A neurodegenerative disease is caused by the progressive loss of structure or function of neurons, in the process known as neurodegeneration. Such neuronal damage may ultimately involve cell death. Neurodegenerative diseases include amyotrophi ...
. This suggests that DNA is incapable of reentering the repair pathway without additional back-up machinery to correct for ligase errors.
With the structure of DNA being well known and many of the components necessary for its manipulation, repair, and usage becoming identified and characterized, researchers are beginning to look into the development of nanoscopic machinery that would be incorporated into a living organism that would possess the ability to treat diseases, fight cancer, and release medications based on a biological stimulus provided by the organism to the nanosocpic machinery. DNA ligase would most likely have to be incorporated into such a machine.
References
Further reading
* * * * * * * * * * * * * * * * * *External links
* {{Portal bar, Biology, border=no EC 6.5.1